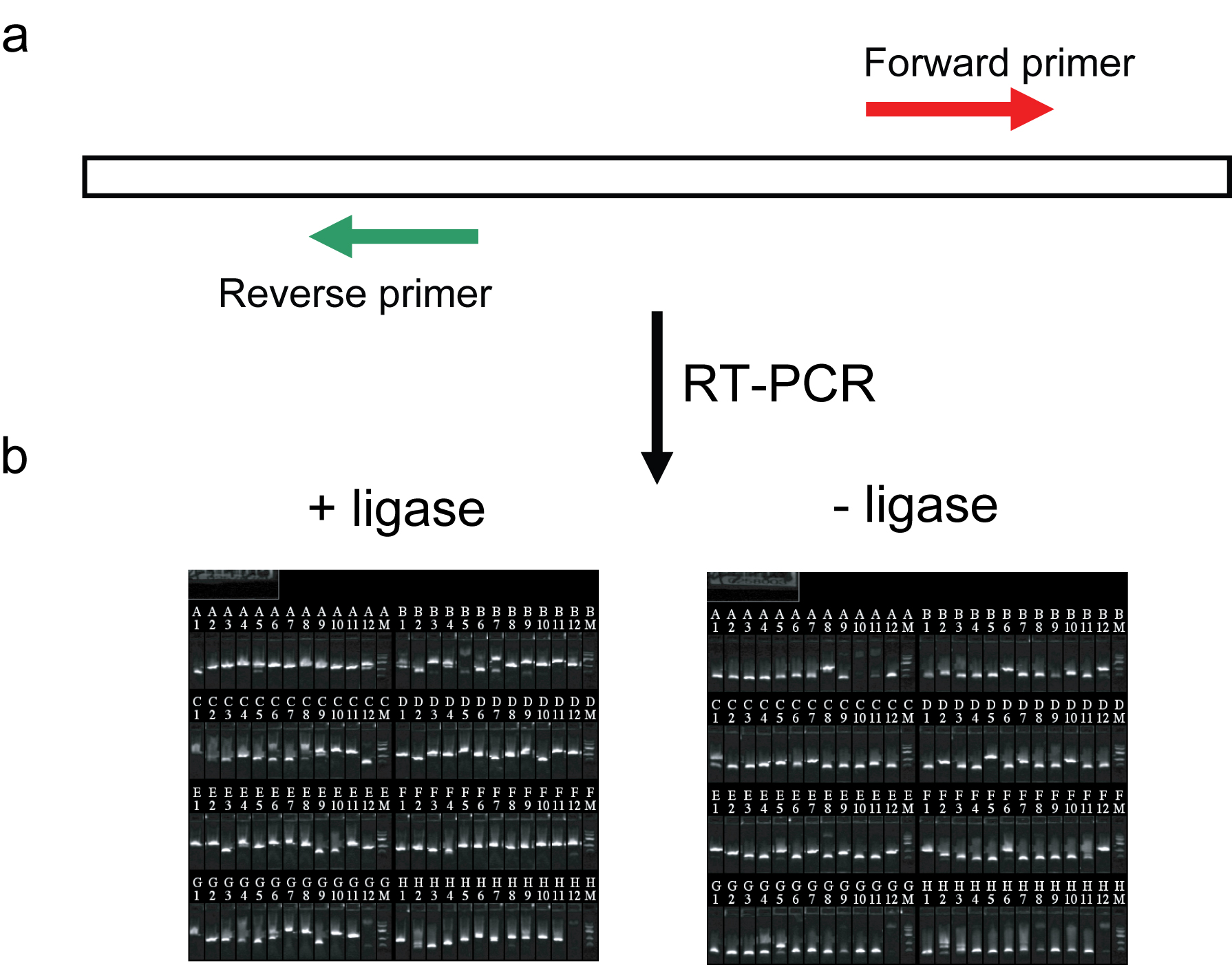
Circular RNAs of protein coding genes have been observed occasionally (Capel et al., 1993). Biological properties of such transcripts have not been deeply explored, nor have their occurrence in C. elegans been described. As the functional properties of these transcripts can be effectively evaluated by future RNAi knockdown experiments in the worm, we investigated whether circularized RNAs are expressed. Our approach (Panel a) entails amplification of reverse transcribed RNA using primers in which the "reverse" primer matches sequences upstream of the "forward" primer on a transcript. With such primer configurations, a reverse transcribed product made from a linear transcript cannot be amplified, but reverse transcribed product generated from circular transcripts supports amplification. We carried out RT-PCR on total RNA isolated from N2 worms targeting a set of 94 SL1 positive control transcripts we had recently studied by RACE (Salehi-Ashtiani et al., 2009). Most transcript models so tested yielded a band in RT-PCR reaction without addition of RNA ligase (Panel b, "-ligase"), suggesting that circular RNA formation is common in vivo. The obtained PCR products were cloned as minipools, then end-sequenced from both directions. Alignment of the obtained sequences to the genome revealed “circular junctions” in 37 of 94 transcript models examined. These sequences were spliced, although no SL or poly(A) sequences were detected, suggesting that these transcripts had circularized before post-transcriptional processing, or had lost these modifications prior to circularization. As a control, RNA ligase 1 was added to RNA before reverse transcription to generate circularized RNA. Cloning and sequencing of RT-PCR products (Panel b, "+ligase") made from these RNAs showed that unlike the no-ligase RNAs, SL and/or poly(A) sequences were frequently observed at the junctions, indicating that the absence of these modifications in the no-ligase experiments was not due to technical limitations. As a possible biological function, we note that translation of circular transcripts (e.g., through an internal ribosome entry site), can expand the genome coding potential by juxtaposing exons in a way not obtainable through alternative splicing of linear transcripts.
Figures
References
Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, and Lovell-Badge R. (1993). Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73, 1019-1030. 
Salehi-Ashtiani K, Lin C, Hao T, Shen Y, Szeto D, Yang X, Ghamsari L, et al. (2009). Large-scale RACE approach for proactive experimental definition of C. elegans ORFeome. Genome Res. [Epub ahead of print].