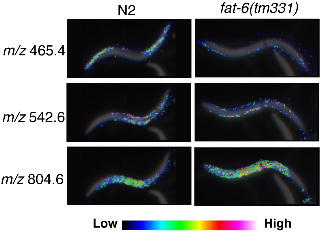
Imaging mass spectrometry (IMS) is a technique based on two-dimensional mass spectrometry to visualize the spatial distribution of biomolecules, which does not need separation, purification or labeling of target molecules (Kimura et al., 2009). Using tissue sections of mammals we, and others, have succeeded to visualize the distribution of peptides, lipids and other small metabolic molecules. Here, we report the first application of IMS to C. elegans. MALDI (matrix-assisted laser desorption ionization) imaging has revealed that numerous biomolecules are localized specifically in the limited regions. The resolutions of the images obtained by IMS are high enough to analyze the biomolecules even in small C. elegans (Fig.).
We compared wild type (N2) and fat-6(tm331) mutants, which lack a Δ9 fatty acyl desaturase, and observed a dramatic difference in expression level and localization of biomolecules (Fig.). We have not studied the molecular identity of these molecules yet, however, MS/MS analysis can give us detailed nature of the molecular structure. We hope that IMS could bring innovation to the C. elegans research community as a new microscopy to observe worms and a convenient tool for mutant screening.
Figures
References
Kimura Y, Tsutsumi K, Sugiura Y and Setou M. (2009). Medical molecular morphology with imaging mass spectrometry. Med. Mol. Morphol. 42, 133-137.