A barrier to rigorous tests for the role of mitochondrial dysfunction in aging processes has been the lack of model systems with relevant, naturally occurring mitochondrial genetic variation. Toward the goal of developing such a model, we studied life history, metabolic, and aging phenotypes in natural and experimental populations of Caenorhabditis briggsae harboring different levels of a heteroplasmic mitochondrial ND5 deletion recently discovered to segregate among many wild C. briggsae populations (Howe and Denver, 2008). The normal product of ND5 is a central component of complex I of the mitochondrial electron transport chain and integral to cellular energy metabolism.
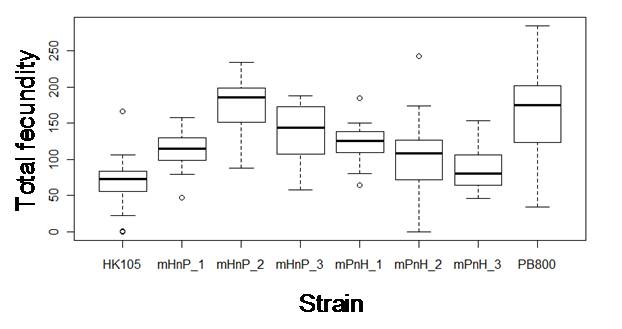
Natural high-ND5 deletion C. briggsae isolates have decreased total fecundity (Howe and Denver, 2008). We found that heteroplasmic C. briggsae also have reduced pharyngeal pumping rates throughout adulthood and grow faster and larger as larvae, but slower as adults, compared to zero-deletion isolates. Hybrid strains were constructed by reciprocally crossing pairs of high- and low-heteroplasmy populations in order to isolate the mitochondrial genome of one population onto the nuclear genetic background of another population. F10 individuals were assayed for the above traits with the expectation that hybrid means would match those of the mitochondrial source population if ND5 deletion heteroplasmy level is responsible for the phenotypic variation among C. briggsae isolates. However, C. briggsae hybrid means often differed from those of maternal lineage and from each other for all traits (e.g., Fig. 1). Because coordination between nuclear and mitochondrial gene products is required for optimal energy metabolism, our results may indicate cytonuclear conflict and heterosis in hybrid C. briggsae, but paternal transmission of mitochondria has yet to be eliminated as a potential cause of these results.
Figures
References
Howe DK, and Denver DR. (2008). Muller's ratchet and compensatory mutation in Caenorhabditis briggsae mitochondrial genome evolution. BMC Evol. Biol. 8, 62. 




