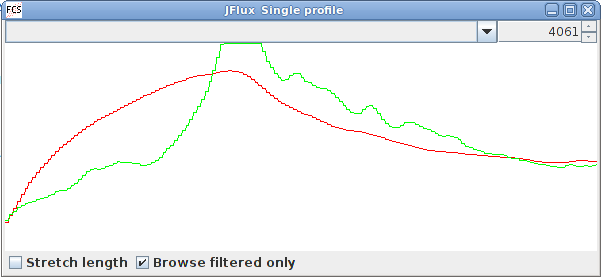
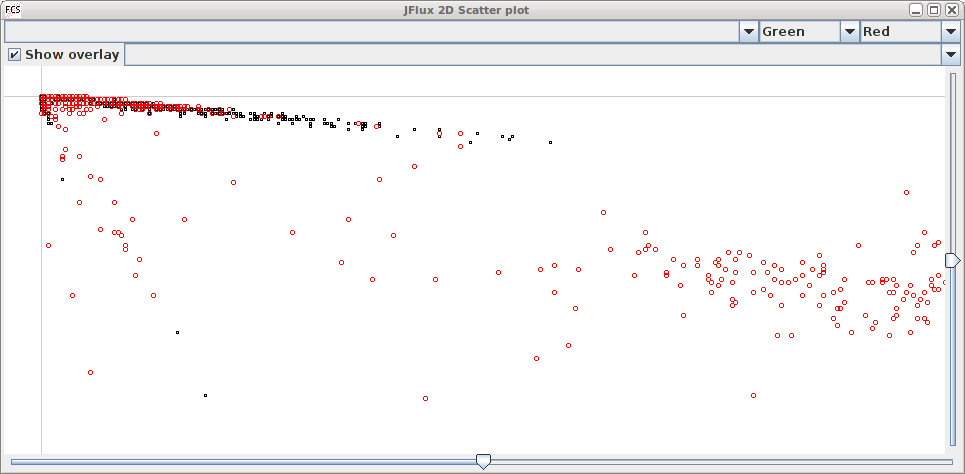
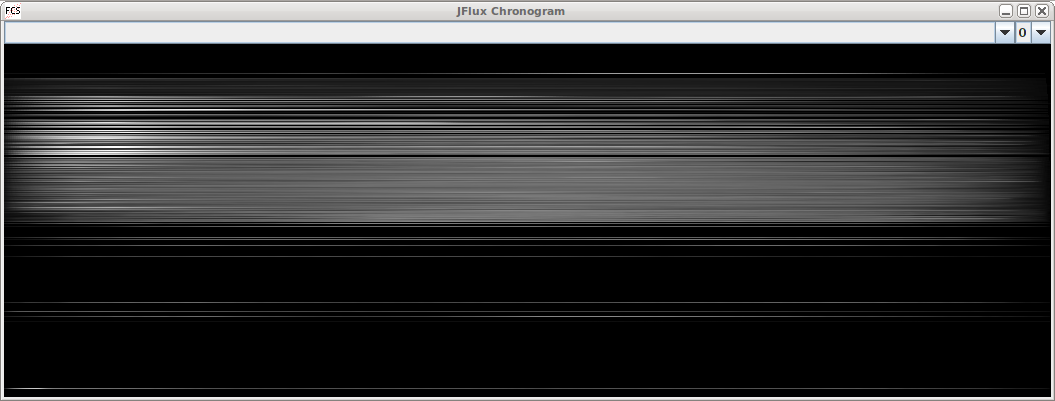
We recently acquired a COPAS worm sorter and found that the analysis software included could not do what we needed. For this reason, we began developing a framework tailored to analyze the data. So far we have developed the functionality to produce chronograms (Dupuy et al., 2007) and to compare subregions statistically. The scatter plots produced are easy to navigate and can be overlaid to compare different runs.
The software is open source, enabling anyone to extend or modify it to fit their purpose. It is split into two parts: a library that can be used within your own code (e.g. for high-throughput analysis) and a graphical user interface for manual visualization and analysis of the data. All code is written in Java.
The code is already available at jflux.sourceforge.net, but the software is still in early development. We hope that other COPAS users are interested in contributing to this project. Having an extensible platform will allow us to incorporate new analysis methods to make the most out of the data.
Figures
References
Dupuy D, Bertin N, Hidalgo CA, Venkatesan K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al. (2007). Genome-scale analysis of in vivo spatiotemporal promoter activity in Caenorhabditis elegans. Nature Biotechnology 25, 663-668.