Social interactions, learning experiences and responses to wide-ranging environmental stimuli occur through nervous cells via synapses. Several observations have suggested that the alteration of neuron connections during nervous system development could represent the root of many cases of autism spectrum disorder. Neuroligins are synaptic cell adhesion proteins that have been shown to be able to induce synaptogenesis. Single missense and frameshift mutations in neuroligin coding genes have been proposed that may lead to autism or mental retardation with complete penetrance (Baudouin and Scheiffele, 2010; Etherton et al., 2011). In C. elegans, nlg-1 is orthologous to mammal neuroligin genes, and it has been shown that nlg-1 mutants are defective in several sensory behaviors and sensitive to oxidative stress (Calahorro et al., 2009; Hunter et al., 2010).
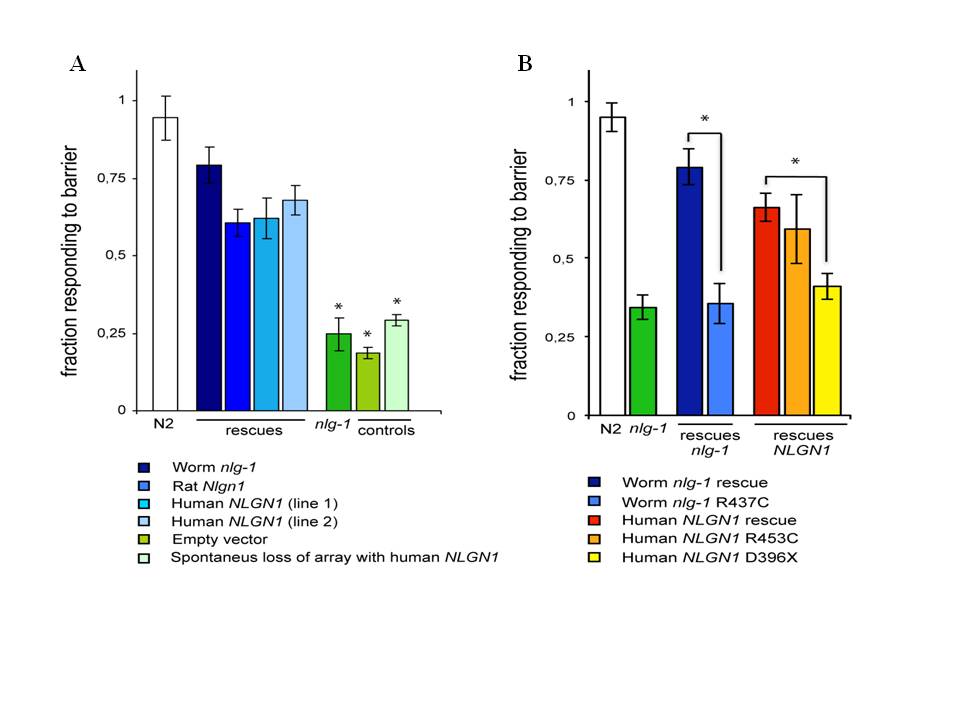
We report here that transgenic expression of rat Nlgn1 and human NLGN1 proteins are functional in C. elegans. The wild type behavior pattern was rescued in nlg-1 deficient mutants by expressing cDNAs from rat Nlgn1 or human NLGN1 genes under C. elegans nlg-1 promoter (Figure 1A). Induced changes Asp396Stop in human NLGN1 or Arg453Cys in worm NLG1 proteins, failed to rescue the wild type phenotype (Figure 1B). These results indicate that mammalian and C. elegans neuroligins seem to be functionally comparable. The results anticipate that the nematode could be useful as an in vivo model for studying specific synapse mechanisms involved in autism. This system will allow the analysis of how mutations in neuroligin genes change phenotypes in different C. elegans genetic backgrounds and the study of their interactions with different environmental factors. It is probable also that this approach could be extended to other genes encoding synaptic proteins implicated in autism spectrum disorder, such as neurexins and shanks.
Table 1. Strains used in this study
| Name | Genotype | Reference/Source |
| N2 | wild type reference | CGCa |
| VC228 | nlg-1 (ok259) X | CGCa |
| CRR1b | nlg-1 (ok259) X | This study |
| CRR100 | nlg-1 (ok259) X; crrEx4 [pPD95.77; pDD04 NeoR (pmyo-2::GFP)] | This study |
| CRR104c | nlg-1 (ok259) X; crrEx4 [pPD95.77 (Pnlg-1::nlg-1); pDD04NeoR (pmyo-2::GFP)] | This study |
| CRR105 | nlg-1 (ok259) X; crrEx5 [pPD95.77 (Pnlg-1::nlg-1-R437C); pDD04NeoR (pmyo-2::GFP)] | This study |
| CRR106d | nlg-1 (ok259) X; crrEx6 [pPD95.77 (Pnlg-1::NLGN1); Pnrx-1::GFP] | This study |
| CRR107 | nlg-1 (ok259) X; crrEx7 [pPD95.77 (Pnlg-1::NLGN1-R453C); pDD04NeoR (pmyo-2::GFP)] | This study |
| CRR108 | nlg-1 (ok259) X; crrEx8 [pPD95.77 (Pnlg-1::NLGN1-D396X); pDD04NeoR (pmyo-2::GFP)] | This study |
| CRR109e | nlg-1 (ok259) X; crrEx9 [pPD95.77 (Pnlg-1::Nlgn1::EGFP); pBCN24NeoR] | This study |
a Caenorhabditis Genetics Center.
b Obtained by outcrossing VC228 strain with N2 six times.
cThe cDNA nlg-1 coding region was obtained from clone yk1657a10, Yuji Kohara, National Institute of Genetics, Mishima, Japan.
dThe cDNA human NLGN1 coding region was obtained from clone KIAA1070 (hj05602), Kazusa DNA Research Institute, Japan.
e Rat Nlgn-1::EGFP was a gift from Dr. Thomas Dresbach, Univ. Göttingen, Germany.
Figures
References
Baudouin S and Scheiffele P. (2010). SnapShot: neuroligin-neurexin complexes. Cell 141, 908-908.e1. 
Calahorro F, Alejandre E and Ruiz-Rubio M. (2009). Osmotic avoidance in Caenorhabditis elegans: synaptic function of two genes, orthologues of human NRXN1 and NLGN1, as candidates for autism. J. Vis. Exp. 34, pii: 1616. 
Etherton MR, Tabuchi K, Sharma M, Ko J and Südhof TC. (2011). An autism-associated point mutation in the neuroligin cytoplasmic tail selectively impairs AMPA receptor-mediated synaptic transmission in hippocampus. EMBO J. 30, doi:10.1038. 
Hunter JW, Mullen GP, McManus JR, Heatherly JM, Duke A and Rand JB. (2010). Neuroligin-deficient mutants of C. elegans have sensory processing deficits and are hypersensitive to oxidative stress and mercury toxicity. Dis. Model. Mech. 3, 366-376. 




