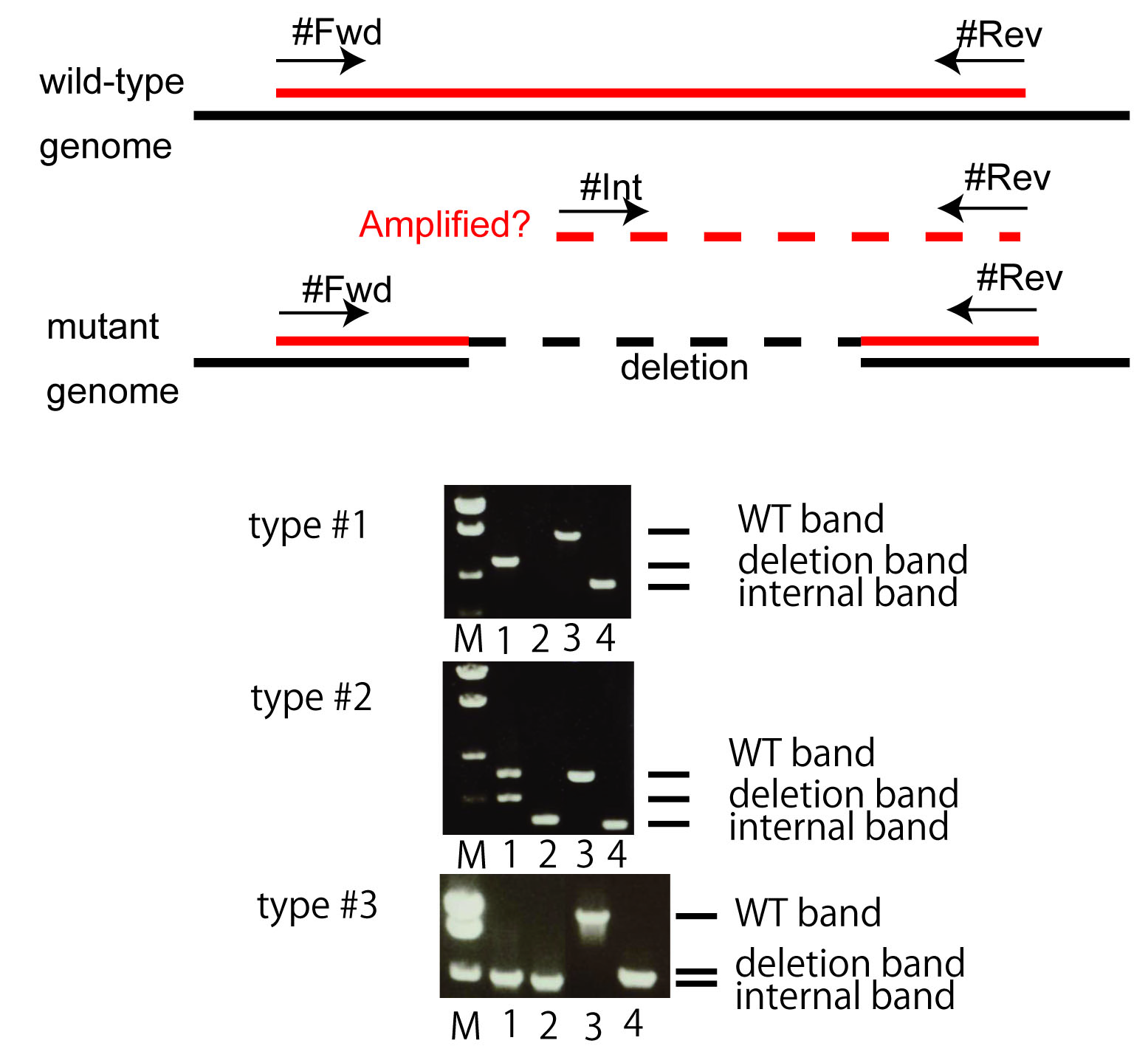
We are isolating and distributing deletion mutants to the community using a TMP-UV method (Gengyo-Ando and Mitani, 2000). To collect high-quality mutants, we used to examine the presence of wild-type DNA sequences when deletion sizes are longer than 1 kb. However, after a time-point, we changed the criteria shorter than before: 500 bp. When deletion sizes are shorter, we can easily identify such a rearrangement as we do "homo-hetero" tests (as seen by WT band in lane 1 in addition to internal band in lane 2 of type #2 mutant in Figure 1). Most of the mutants caused by rearrangements can be excluded this way and not posted publicly. We guess, however, there may be still a small number of such rearrangement artifacts in our mutant collections in earlier tm alleles (before tm2000). We re-examine the rearrangements when necessary and continuously improve the quality of the collection; we do the re-examination when we find strains that turned out to be homozygous viable, which were regarded as homozyogous inviable and did not examine the internal primer test before. We withdraw alleles when we find problems and try to replace them by isolating new alleles. We hope if people find such cases, please let us know.
Figures
References
Gengyo-Ando K and Mitani S. (2000). Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 269, 64-69.





