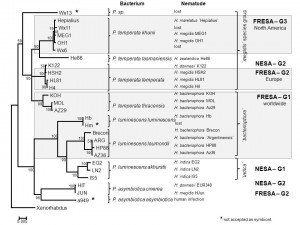
We have recently established an agar based media (ENGM) amenable to use in genetic studies of entomopathogenic nematodes (Fodor et al., WBG 18:2). Herein we present some results confirming the practical usefulness of this media. Three recognized Photorhabdus species (P. asymbiotica, P. temperata and P. luminescens) include subspecies, which, apart from P. asymbiotica ssp. asymbiotica are obligate endosymbionts of different heterorhabditid nematodes, (Peat et al., 2010). We used six different enzymes (AluI, TaqI, MseI, HinfI, RsaI, and DdeI) to establish the PhastSystem PAGE RFLP patterns (as phenotypes) of the intergenic spacer sequences (IGS) within the 16S – 23S rRNA operon of the natural EPB symbionts (Pamjav et al., 1999). Gnotobiological analysis (culturing nematodes on each others’ symbionts and evaluating the results) were carried out on ENGM to delineate partner specificities and host ranges. The study covers majority but not all of the known respective symbiotic complexes. We described Symbiotic Association (SA) Groups within which the symbiotic partners could not (NESA-G) or could (RESA-G1 & 2) reciprocally be exchanged. In one SA group (RESA-G3) the option for symbiotic partner exchange is occasional. Both partners of NESA-G1 are monophyletic (H. indica and P. luminescens ssp. akhurstii strains). All EPN partners in RESA-G1 (H. bacteriophora) and in RESA-G2 (H. megidis North Western European strains) are monophyletic, but the EPB partners are polyphyletic. In RESA-G3 both the EPN (H. megidis, H. marelatus some H. bacteriophora strains) and the EPB partners are polyphyletic. The data of exhibiting gnotobiological promiscuity related to exceptional symbiont exchanges between H. marelata, H. megidis, H. bacteriophora and H. downesii strains suggest that alternative co-evolutionary pathways might be initiated by fixation of a novel natural Heterorhabditis/Photorhabdus symbiotic association. Once an EPN/EPB SA established the evolutionary fate of the bacterial, but the nematode partner became unambiguously determined and canalized. We concluded that the Heterorhabditis/Photorhabdus co-evolution is a “one-way street”.
Figures
References
Pamjav H, Triga D, Buzás Z, Vellai T, Lucskai A, Adams B, Reid AP, Burnell A, Griffin C, Glazer I, Klein MG, and Fodor A. (1999). Novel application of PhastSystem polyacrylamide gel electrophoresis using restriction fragment length polymorphism of internal transcribed spacer patterns of individuals for molecular identification of entomopathogenic nematodes. Electrophoresis 20, 1266-1273. 
Peat SM, Ffrench-Constant RH, Waterfield NR, Marokházi J, Fodor A, and Adams, BJ. (2010). A robust phylogenetic framework for the bacterial genus Photorhabdus and its use in studying the evolution and maintenance of bioluminescence: a case for 16S, gyrB, and glnA. Mol. Phylogenet. Evol. 57, 728–740. 




