Dosage compensation equalizes X chromosome gene expression between the sexes. In Caenorhabditis elegans, the dosage compensation complex (DCC) binds both hermaphrodite X chromosomes and downregulates expression from each two-fold. Documented in a recent paper, we found that repressive H4K20me1 is enriched on hermaphrodite X chromosomes in a sex- and DCC-dependent manner (Wells et al., 2012). H4K20me1 levels are regulated by the conserved methyltransferases SET-1 (PR-SET7 homolog) and SET-4 (SUV4-20 homolog). PR-SET7 is a monomethylase, while SUV4-20 is a di- and tri-methylase, in other systems, and our data support conservation of these roles in C. elegans (Wells et al., 2012). We hypothesized that knockdown of both set-1 and set-4 would cause a derepressed chromatin state in developing worms.
To look for this effect, we employed a modified xol-1 suppression assay (Miller et al., 1988) using a him-8(e1489); xol-1(y9) sex-1(y263) strain in which males die due to the inappropriate dosage compensation allowed by a xol-1 mutation. A mutation in sex-1 sensitizes the worms to dosage compensation defects. Male viability is rescued by depleting factors important for dosage compensation function.
Simultaneous RNAi of multiple factors is known to be inefficient (Shapira and Tan, 2008; Michael Wells, unpublished observations). Several studies have employed sequential RNAi strategies to overcome this issue (Shapira et al., 2006; Checci and Engebrecht, 2011). Using sequential RNAi, it was possible to knock down a non-essential gene (set-4), then an essential gene in the same pathway (set-1), in the rescue assay (Wells et al., 2012). Length of exposure to each RNAi can be varied to obtain the desired level of knockdown by changing growing temperature and when in development worms are transferred.
Two-generation set-1 RNAi treatment led to less than 5% viability in this strain, precluding counts. Simultaneous knockdown with a mixture of set-1 and set-4 RNAi-expressing bacteria showed insignificant male rescue. Two-generation set-4 knockdown led to 4.5% rescue, as compared to less than 1% with empty vector and 54% with dpy-27, a DCC member, RNAi; finally, set-4 followed by set-1 RNAi led to a remarkable 20% rescue of male viability (Wells et al., 2012).
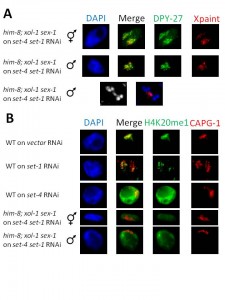
The rescued males grew up to become adults of a smaller size than WT males, but possessed a well-developed tail and a single gonad arm (data not shown). We showed that these rescued males are indeed XO using X chromosome FISH in meiotic anaphase sperm nuclei (Fig. 1A), where the single X lags behind separating autosomes. Male rescue assay results indicate that DCC function is compromised in these animals (Miller et al., 1988); however, we observe strong DCC localization to X (Fig. 1A), suggesting that loss of repression is occurring downstream of DCC loading. set-1 RNAi weakened X enrichment of H4K20me1, while set-4 RNAi led to robust, uniform H4K20me1 staining across the nucleus. Following set-4 set-1 sequential RNAi, H4K20me1 levels in both sexes were low and similar across the entire nucleus (Fig. 1A), possibly due to loss of X enrichment (set-4 RNAi) and greatly reduced monomethylation (set-1 RNAi).
These data indicate that loss of set-1 and set-4 caused a disruption in dosage compensation-mediated repression, downstream of DCC loading, sufficient for males to survive to the adult stage. We have since continued to exploit this robust sequential RNAi method to study other essential factors, and believe that sequential RNAi could be used in many useful contexts.
Figures
References
Checchi P, and Engebrecht J. (2011). Caenorhabditis elegans histone methyltransferase MET-2 shields the male X chromosome from checkpoint machinery and mediates meiotic sex chromosome inactivation. PLoS Genetics 7, e1002267. 
Miller L, Plenefisch J, Casson L, and Meyer B. (1988). xol-1: A gene that controls the male modes of both sex determination and X chromosome dosage compensation in C. elegans. Cell 55, 167-183. 
Shapira M, Hamlin B, Rong J, Chen K, Ronen M, and Tan M. (2006). A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc. Natl. Acad. Sci. USA 103, 14,086-14,091. 
Shapira, M, and Tan M. (2008). Genetic analysis of Caenorhabditis elegans innate immunity. Methods Mol. Bio. 415, 429-442. 
Wells M, Snyder M, Custer L, and Csankovszki G. (2012). Caenorhabditis elegans dosage compensation regulates histone H4 chromatin state on X chromosomes. Mol. Cell. Biol. 32, 1710-1719.