Of the 1090 somatic cells generated during development of C. elegans hermaphrodites, 131 undergo programmed cell death. Transcription factors that regulate cell-specific apoptosis in C. elegans have been associated with cancer (Reviewed in Potts et al., 2011). We aimed to identify novel transcription factors that regulate programmed cell death in C. elegans by performing an RNAi screen against transcription factors in a sensitized cell death defective background. A homolog of human leukemia associated AF10, zfp-1, was identified as a hit, as it was found to promote cell death in C. elegans. Our initial characterization of the programmed cell death defects of zfp-1 mutants is described here.
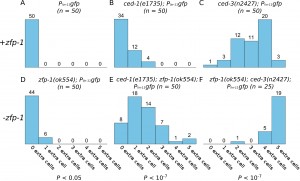
We performed an RNAi screen of 387 transcription factors and 263 chromatin remodeling factors using ced-3(n2427); Plin-11gfp worms to identify genes that regulate cell death in the ventral nerve cord. The Plin-11gfp reporter expresses GFP in the VC motor neurons of the ventral nerve cord, and provides a quick and reliable assay for identifying regulators of cell death (Reddien et al., 2001). In Plin-11gfp (otherwise genetically wild-type) worms (Figure 1A), four cells in the ventral nerve cord are clearly visible under a dissecting fluorescence microscope; two of the six VC neurons are obscured by fluorescence in vulval cells, which also express the reporter. In worms with mutations that result in a strong cell death defect, Plin-11gfp is expressed in five additional cells in the ventral nerve cord. These additional cells are the lineal equivalents of ventral neurons in the anterior and posterior ventral nerve cord, which undergo programmed cell death in wild type animals. In mutants with partial defects in programmed cell death, an intermediate number of additional cells survive and express Plin-11gfp. Worms defective in engulfment of cell corpses (ced-1(e1735)) or with a partial loss of function mutation in the caspase (ced-3(n2427)) have an intermediate number of additional cells that survive and express Plin-11gfp (Figure 1B, 1C). The sensitized ced-1(e1735) and ced-3(n2427) genetic backgrounds can enhance detection of weak cell death defects.
The strongest cell survival phenotype observed, both in the ced-3(n2427) background and in a secondary screen using ced-1(e1735), was in animals fed a zfp-1(RNAi) construct (data not shown). The zfp-1(ok554) deletion allele removes the leucine-zipper domain, rendering the protein non-functional (Cui et al., 2006). When zfp-1(ok554) is crossed into the Plin-11gfp reporter, there is a small, but significant, increase in the number of surviving ventral nerve cells (Figure 1D). The weak effect of zfp-1(ok554) in an otherwise wild type reporter background is enhanced in ced-1 and ced-3 mutants with partial defects in programmed cell death (Figure 1E, 1F). Other transcription factors, such as lin-35, can enhance a weak cell death defect (Reddien et al., 2007), and zfp-1 regulates a similar subset of genes (Grishok et al., 2008). It is possible that zfp-1 transcriptionally regulates cell death in a manner similar to that of lin-35. Thus, knockdown of zfp-1 might elicit a cell death phenotype as a result of its activity in transcriptional regulation.
Figures
References
Cui M, Kim EB, Han M. (2006) Diverse chromatin remodeling genes antagonize the Rb-involved SynMuv pathways in C. elegans. PLoS Genet 2, e74. 
Grishok A, Hoersch S, Sharp PA. (2008) RNA interference and retinoblastoma-related genes are required for repression of endogenous siRNA targets in Caenorhabditis elegans. Proc Natl Acad Sci U S A 105, 20386–20391. 
Potts M B, Cameron S. (2011) Cell lineage and cell death: Caenorhabditis elegans and cancer research. Nat Rev Cancer 11, 50–58. 
Reddien PW, Andersen EC, Huang MC, Horvitz HR. (2007) DPL-1 DP, LIN-35 Rb and EFL-1 E2F act with the MCD-1 zinc-finger protein to promote programmed cell death in Caenorhabditis elegans. Genetics 175, 1719–1733. 
Reddien PW, Cameron S, Horvitz HR. (2001) Phagocytosis promotes programmed cell death in C. elegans. Nature 412, 198–202.