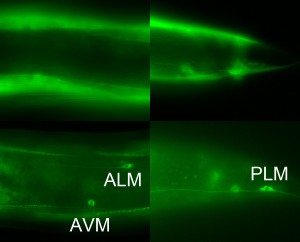
Promoter or translational gfp fusions are commonly used to visualize gene expression patterns and protein localizations. The labeling of neurons by GFP and other fluorescent proteins, however, can be obscured by strong expression in adjacent tissues, such as the body-wall muscle, the hypodermis and the gut. One can circumvent this problem by only marking nuclei using GFP fused to a nuclear localization signal, but this method loses information about the shape of the cell, which facilitates cell identification.
We have devised a simple method to aid identification of neurons expressing gfp by reducing gfp expression in non-neuronal tissues. Feeding RNAi does not effectively knockdown gene expression in neurons (Timmons et al., 2001), possibly because neurons generally do not express SID-1, a dsRNA transporter required for systemic RNAi (Winston et al., 2002; Calixto et al., 2010). Therefore, feeding RNAi against gfp in animals wild-type for sid-1 should eliminate non-neuronal expression of gfp. Figure 1 shows expression of unc-112p::unc-112::gfp in animals fed bacteria expressing dsRNA against gfp (bottom) and those fed non-RNAi bacteria (top). UNC-112, a dense body component homologous to the human kindlins (Rogalski et al., 2000), is active in body wall muscles (Rogalski et al., 2000), and touch receptor neurons (Calixto et al., 2010). The muscle expression is knocked down without eliminating GFP in the TRNs. Our method therefore provides a convenient means to identify gfp expression in neurons.
Figures
References
Calixto A, Chelur D, Topalidou I, Chen X, Chalfie M. (2010) Enhanced neuronal RNAi in C. elegans using SID-1. Nat Methods 7, 554-559. 
Rogalski TM, Mullen GP, Gilbert MM, Williams BD, Moerman DG. (2000) The unc-112 gene in Caenorhabditis elegans encodes a novel component of cell matrix adhesion structures required for integrin localization in the muscle cell membrane. J Cell Biol 150, 253-264. 
Timmons L, Court DL, Fire A. (2001) Ingestion of bacterially expressed ds RNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103-112. 
Winston WM, Molodowitch C, Hunter CP. (2002) Systemic RNAi in C. elegans requires the putative transmembrane pretein SID-1. Science 295, 2456-2459.