The C. elegans genome encodes a large family of putative insulin-like peptides comprising 40 genes (Pierce et al., 2001; Li et al. 2003). In spite of great interest in the insulin-like pathway, given its function in dauer formation, aging, and L1 arrest, the function of most insulin-like peptides remains uncharacterized. There is evidence, however, that they function as either agonists or antagonists of the insulin-like receptor DAF-2, promoting development or arrest, respectively (Pierce et al., 2001). The lack of loss-of-function phenotypes for most insulin-like peptides points towards redundancy, although to what extent remains unknown. Recent findings of Ritter et al. indicate that no single insulin is the sole agonist or antagonist for coordinating dauer formation through the DAF-2 receptor and that there may be complex patterns of redundancies between multiple insulins (Ritter et al., 2013).
We performed a microarray analysis comparing the gene expression between 24h starved L3 stage N2 and fully fed animals of the same age (Figure 1). The starved animals displayed a delay in growth and were still the size of L3 worms, whereas the fully fed animals developed normally to late L4 stage at the time of sampling. 37 of the insulin-like peptide genes (ins) were represented on the Agilent C. elegans Gene Expression Microarrays (design ID 020186).
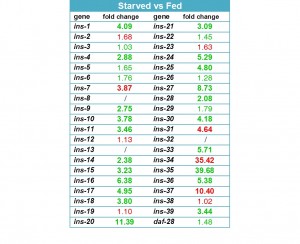
25 of the 40 known ins genes were differentially expressed more than twofold in the starved animals (Figure 2). Remarkably, 21 insulins are upregulated under L3 starvation while only 4 are downregulated. The strongest increase in expression was observed for ins-35 (39.7 fold), ins-20 (11.4 fold), ins-16 (6.4 fold) and ins-33 (5.7 fold); the strongest decrease in ins-34 (35.4 fold), ins-37 (10.4 fold), ins-31 (4.6 fold) and ins-7 (3.9 fold). The high number of differentially regulated insulins seems to favor of a possible functional redundancy between these peptides, given that DAF-2 is the likely receptor for most of them. The strong change in transcript level of many of these peptides is indicative of their importance with regard to starvation survival.
INS-4, INS-6, INS-7 and DAF-28 have been suggested to be DAF-2 agonists (Pierce et al., 2001; Hua et al., 2003; Murphy et al. 2003), while INS-1, INS-17, INS-18, INS-33 and INS-35 represent antagonists (Pierce et al., 2001; Liu et al., 2004). Our data is consistent with a possible agonistic role for INS-7 (downregulated) and antagonistic role for INS-1, INS-17, INS-18, INS-33 and INS-35 (upregulated) with regard to L3 starvation and its resulting developmental effects. INS-4 and DAF-28 transcript levels, however, were not differentially regulated, and INS-6 was upregulated, indicating it might act as an antagonist under the condition of L3 starvation. The data also suggests possible new agonistic and antagonistic roles for many other insulin-like peptides, for example: INS-31, INS-34 and INS-37 as agonists, inhibiting developmental arrest; and INS-16, INS-20, INS-24, INS-25, INS-30 and INS-36 as antagonists, stimulating developmental arrest. A functional role for these insulins was not described before and might be specific to the condition of L3 starvation.
Different groups of insulins might therefore act differently depending on the physiological and environmental conditions, a concept that was also recently described in the ‘‘block design’’ by Ritter et al. (2013).
Figures
References
Hua QX, Nakagawa SH, Wilken J, Ramos RR, Jia W, Bass J, and Weiss MA. (2003). A divergent INS protein in Caenorhabditis elegans structurally resembles human insulin and activates the human insulin receptor. Genes Dev. 17, 826-831. 
Li W, Kennedy SG, and Ruvkun G. (2003). daf-28 encodes a C elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 17, 844-858. 
Liu T, Zimmerman KK, and Patterson GI. (2004). Regulation of signaling genes by TGFbeta during entry into dauer diapause in C. elegans. BMC Dev. Biol. 4, 11. 
Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, and Kenyon C. (2003). Genes that act downstream of DAF16 to influence the lifespan of Caenorhabditis elegans. Nature 424, 277-283. 
Pierce SB, Costa M, Wisotzkey R, et al. (2001). Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 15, 672-686. 
Ritter AD, Shen Y, Fuxman Bass J, et al. (2013). Complex expression dynamics and robustness in C. elegans insulin networks. Genome Res. 23, 954-965. 




