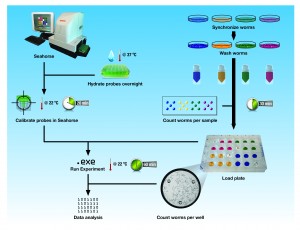
This is a protocol for the determination of oxygen consumption rate (OCR) in living C. elegans using the Seahorse apparatus with 24-well plates. OCR provides a measurement of respiration, which can be altered by genetic or pharmacologic manipulation of the mitochondrial respiratory chain. Although the Seahorse was originally designed for use with ex vivo cells, it can readily be used in vivo with living C. elegans by following our simple procedure,2 described here for the analysis of four samples side-by-side.
Synchronize each of the four samples of worms. We allow 200 adults to lay eggs for 4 h on a 10 cm plate by picking 200 adults to a 10 cm plate, kill the adults, then study the F1s as day 1 adults. Program the Seahorse Bioscience XF24 Extracellular Flux Analyzer with the following protocol: 1- calibrate probes; 2- loop 10 times; 3- mix 2 min; 4- time delay 2 min; 5- measure 2 min; 6- loop end. Furthermore, the internal heater should be switched off to allow the instrument to be close to room temperature. Hydrate the probes in the extracellular flux assay kit by adding 1 ml of Seahorse calibrant solution to each well and incubating overnight at 37 oC . Load this into the Seahorse on the day of the experiment for calibration.
Rinse the worms off the plate and wash three times using gravity separation in M9 and resuspend each washed sample in 2 ml. Get a rough estimate of the concentration of worms by pipetting 5 drops of 20 µl each per sample onto an empty dish and counting under the dissecting scope. There should be about 2 worms per drop. In the Seahorse assay plate, place 500 µl M9 into four blank wells. At least 30 min after the worms were first rinsed, pipet the four worm samples into a total of 500 ul M9 in each of the other wells with between 25 and 150 worms per well and five replicates of each sample. We use a low retention tip cut to create a wider opening, and found that picking worms into the wells with a platinum wire leads to unstable results.
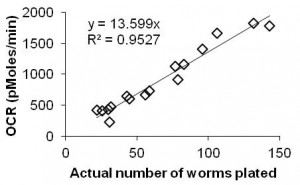
When prompted by the instrument, replace the calibration plate with the worm plate. At the end of the run, count the actual number of worms per well; for this purpose, we capture images on a camera-fitted dissecting scope. Average the background-corrected OCR (pMol O2/min) across the 10 time-points for each well, then normalize the OCR to the actual number of worms in each well. Calculate the average and standard deviation of OCR per worm across the five replicates for each sample. A moderate amount of variation is expected between time-points, wells, plates, and days: we therefore use a minimum of 10 time points of 5 wells, and on at least 2 plates and/or days, to get a meaningful result.
1 For a more detailed protocol with additional tips, please contact Beverley at Beverley.M.Dancy@gmail.com.
2 We acknowledge other labs using Seahorse for C. elegans who provided some tips when we began this work, including Laurent Mouchiroud in Johan Auwerx’s lab at Ecole Polytechnique Fédérale de Lausanne, David Raizen at University of Pennsylvania, and Paul Brookes at University of Rochester Medical Center.
3 We thank José Antonio Mari (ja_mari@yahoo.com) for putting together the graphics in Figure 1.
Figures