Neuropeptides modulate neural circuits driving adaptive behaviors and key physiological processes such as reproductive behavior, metabolism, feeding, locomotion, and experience-based learning. With over 250 predicted sequences, bioactive peptides are by far the largest and most diverse group of neuromodulatory substances in the C. elegans nervous system (Li and Kim, 2010; Bargmann 2012,). Most of them are thought to function through the activation of G protein-coupled receptors (GPCRs) belonging to the Rhodopsin and Secretin classes. The C. elegans genome encodes at least 120 genes for neuropeptide precursors (FLP, NLP, and INS families), and recent studies have estimated a similar number of about 100 to 150 peptide GPCR genes (Frooninckx et al., 2012; Hobert, 2013). Although peptide signaling has undoubtedly diversified in the nematode lineage, many neuropeptidergic signaling systems share homology to vertebrate and protostomian systems (Mirabeau and Joly, 2013). Growing evidence implicates neuropeptides in many, if not all, behaviors in C. elegans. In contrast, only 23 C. elegans peptide GPCRs (isoforms included) have been deorphanized so far (Frooninckx et al., 2012), leaving more than 80 percent of all putative peptide receptors with unknown ligands.
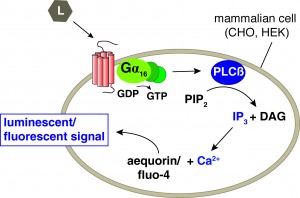
Knowledge on functionally active neuropeptide-receptor couples and GPCR affinity is crucial to further expand our understanding of how neuropeptides function and modulate neural circuits. We are therefore undertaking a large-scale deorphanization initiative that aims at matching all predicted peptide GPCRs to their cognate neuropeptide ligand(s). The Peptide-GPCR project builds further on our combined reverse pharmacology approach (Figure 1), which in the past has enabled us to uncover several C. elegans neuropeptide systems – including cholecystokinin, pigment dispersing factor, and vasopressin/oxytocin related signaling, among others (Beets et al., 2012; and reviewed in Frooninckx et al., 2012). Both knowledge on natural peptides in tissue extracts and in silico peptide predictions are used to maximize the chances to identify a receptor’s cognate ligand(s). Through this combined approach, GPCRs are challenged with a collection of more than 260 peptides belonging to the established FLP and NLP families. The in vitro calcium mobilization strategy allows screening of these hundreds of peptides on all putative peptide GPCRs in a high-throughput manner.
The deorphanization of predicted C. elegans peptide GPCRs should provide a basis for furthering our understanding of nematode neuropeptide signaling and modulation. We aim to share progress on newly identified peptide-GPCR couples to the worm community, as we move down the random list of peptide GPCRs in the course of this project. We are working on a project’s website through which you can already help us prioritize peptides or receptor candidates. Community members are invited to steer the project’s progression via http://worm.peptide-gpcr.org.
Figures
References
Bargmann CI. (2012). Beyond the connectome: how neuromodulators shape neural circuits. Bioessays 34, 458-465. 
Beets I, Janssen T, Meelkop E, Temmerman L, Suetens N, Rademakers S, Jansen G, and Schoofs L. (2012). Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science 338, 543-545. 
Frooninckx L, Van Rompay L, Temmerman L, Van Sinay E, Beets I, Janssen T, Husson SJ, and Schoofs L. (2012). Neuropeptide GPCRs in C. elegans. Front. Endocrinol. (Lausanne) 3: 167, 1-18. 
Hobert O. Neurogenesis in the nematode Caenorhabditis elegans (October 4, 2010), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.12.2, http://54.165.141.74/. 
Li C and Kim K. (2010). Neuropeptide gene families in Caenorhabditis elegans. Adv. Exp. Med. Biol. 692, 98-137. 
Mirabeau O and Joly JS. (2013). Molecular evolution of peptidergic signaling systems in bilaterians. Proc. Natl. Acad. Sci. U. S. A. 110, E2028-2037. 




