 Embryo series courtesy of Einhard Schierenberg
Embryo series courtesy of Einhard SchierenbergAbstract
Protein kinases are one of the largest and most influential of gene families: constituting some 2% of the proteome, they regulate almost all biochemical pathways and may phosphorylate up to 30% of the proteome. Bioinformatics and comparative genomics were used to determine the C. elegans kinome and put it in evolutionary and functional context. Kinases are deeply conserved in evolution, and the worm has family homologs for over 80% of the human kinome. Almost half of the 438 worm kinases are members of worm-specific or worm-expanded families. Such radiations include genes involved in spermatogenesis, chemosensation, Wnt signaling and FGF receptor-like kinases. The C. briggsae kinome is largely similar apart from the expanded classes, showing that such expansions are evolutionarily recent.
Copyright: © 2005 Gerard Manning. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Protein kinases constitute one of the largest and most important of protein families, accounting for ~2% of genes in a variety of eukaryotic genomes. By phosphorylating substrate proteins, kinases modify the activity, location and affinities of up to 30% of all cellular proteins, and direct most cellular processes, particularly in signal transduction and co-ordination of complex pathways. Many of these pathways are highly conserved, and 53 distinct kinase functions and subfamilies appear to have been conserved between yeasts, nematodes, insects and vertebrates, with a further 91 subfamilies of kinases being seen throughout metazoan genomes. This makes kinase signaling particularly amenable to comparative studies, and kinase activity a particularly good readout of the physiological state of any cell.
This chapter will introduce the diversity of kinases in C. elegans, and compare them to those of fungi and other metazoans, as well as to preliminary results from analysis of the C. briggsae kinome.
Most protein kinases share a common ePK (eukaryotic protein kinase) catalytic domain, and can be identified by sequence similarity with Blast or profile hidden Markov models (HMMs). The remaining atypical protein kinases (aPK) belong to several families, some of which have structural, but not sequence similarity to ePKs. We used ePK and aPK HMMs, and Blast/psi-Blast with divergent kinase sequences, to identify protein kinase sequences in C. elegans genomic and expressed sequences (Manning et al., 2002; Plowman et al., 1999). We identified 438 protein kinase genes, including 20 atypical kinases, and an additional 25 kinase fragments or pseudogenes. All sequences and supporting data are available at http://kinase.com, and all but 8 sequences are now identical to wormpep (v. 141) sequences.
To put worm kinases into an evolutionary and functional context, we compared them with the distant kinomes of human, fly, and yeast. At these distances, 1:1 orthology is rare, so we classified each kinase into a hierarchy of groups, families, and sometimes subfamilies (Manning et al., 2002a; Manning et al., 2002b). The classification is based on sequence similarity within the kinase domain, the presence of additional domains, known biological functions, and conservation across divergent genomes. Across the four kinomes, there are 10 groups, 143 families and 212 subfamilies. The classification of each worm kinase is given in Appendix A.
Since kinases perform such a variety of distinct basic cellular functions, it is not surprising to see that 53 subfamilies and functions are present in all four kinomes (Figure 1). A further 91 subfamilies were found in all three metazoan kinomes, including the tyrosine kinase (TK) group and the TKL group, which mediate much of the complexity of intercellular signal transduction. The gain and loss of kinase functions and subfamilies in each evolutionary lineage is also seen. In general, the data support the coelomate clade, where insects are more closely related to vertebrates than to nematodes, rather than the ecdysozoa clade, which groups insects and nematodes together.
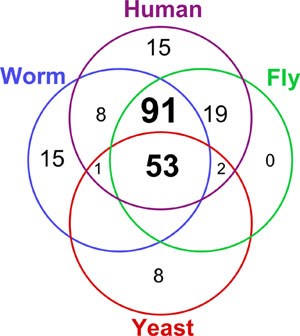
Figure 1. Distribution of 212 kinase subfamilies throughout four kinomes: the yeast Saccharomyces cerevisiae, the nematode worm Caenorhabditis elegans, the fruit fly Drosophila melanogaster and human.
Nematodes share 153 subfamilies with human, providing close homologs for 81% (419/518) of all human kinases. 6 families appear to have been lost from nematodes, based on their presence in fly, human and more basal organisms (Table 1), and several new families have been invented within the coelomate lineage, whose functions predominantly map to immunity/angiogenesis, neurobiology, cell cycle and morphogenesis. There are 13 such human-specific families, and 16 shared between fly and human.
Table 1. Kinases lost in worm, or gained in fly or human kinomes.
| Group | Family | Subfamily | Fly count | Human count | Function | Notes |
|---|---|---|---|---|---|---|
| Secondarily lost from worm | ||||||
| Atypical | G11 | 1 | 1 | Unknown | Also in yeast and plants | |
| CAMK | CAMKL | PASK | 1 | 1 | Glucose sensor | Also in yeast |
| Other | TTK | 1 | 1 | Cell cycle | Also in yeast and plants | |
| Atypical | PIKK | DNAPK | 1 | 1 | DNA repair | Also in Dictyostelium |
| Atypical | TIF | 1 | 3 | Transcriptional control | Also in Dictyostelium. NHR co-factor | |
| TK | CCK4 | 1 | 1 | Neuronal; cell growth | Also in Hydra. Neuronal pathfinding; cancer | |
| Fly + Human | ||||||
| CAMK | Trbl | 1 | 3 | Cell cycle | ||
| CMGC | CDK | CDK10 | 1 | 1 | Cell cycle? | |
| Other | IKK | 2 | 4 | Immunity | NFkb signaling | |
| Other | MOS | 1 | 1 | Cell cycle | Meiosis | |
| Other | SLOB | 2 | 1 | Neuronal | Synaptic transmission | |
| Other | TOPK | 1 | 1 | Cell cycle | ||
| STE | Ste20 | NinaC | 1 | 2 | Neuronal | Phototransduction |
| TK | Jak | 1 | 4 | Immunity | Cytokine signaling | |
| TK | Musk | 1 | 1 | Neuronal | Synaptic transmission | |
| TK | PDGFR/ VEGFR | 2 | 8 | Angiogenesis; morphogenesis; immunity | ||
| TK | Ret | 1 | 1 | Immunity, development | Growth factor receptor | |
| TK | Sev | 1 | 1 | Morphogenesis | ||
| TK | Syk | 1 | 2 | Immunity; morphogenesis | ||
| TK | Tec | 1 | 5 | Immunity; morphogenesis | ||
| TKL | LISK | LIMK | 1 | 2 | Cytoskeletal | |
| TKL | LISK | TESK | 1 | 2 | Testis development | |
| Human | ||||||
| Atypical | Alpha | ChaK | 0 | 2 | Neuronal | Human adds kinase to metazoan-wide channel |
| Atypical | BCR | 0 | 1 | Cell growth | Ras/MAPK growth factor responses | |
| Atypical | FAST | 0 | 1 | Apoptosis | ||
| Atypical | H11 | 0 | 1 | Apoptosis? | ||
| CAMK | CAMKL | HUNK | 0 | 1 | Development | Mammary gland development |
| CAMK | Trio | 0 | 6 | Muscle | Human adds kinase to conserved protein | |
| Other | NKF3 | 0 | 2 | Unknown | ||
| Other | NKF4 | 0 | 2 | Cytoskeletal | ||
| Other | NKF5 | 0 | 2 | Testis development? | ||
| TK | Axl | 0 | 3 | Cell growth; adhesion | ||
| TK | Lmr | 0 | 3 | Cell growth? | ||
| TK | Tie | 0 | 2 | Angiogenesis | ||
| TKL | RIPK | 0 | 5 | Immunity | ||
On the other hand, the worm shares eight subfamilies with human which are absent from Drosophila (Table 2). These include two receptor tyrosine kinase families, an atypical elongation factor 2 kinase (eEF2K), several members of the CAMK group (MELK, PSK, PIM) and the HH498 subfamily of Mixed Lineage Kinases (MLK). In some but not all cases, the fly genome has related genes that may fulfill a similar function. SGK, eEF2K and HH498 are found in Dictyostelium, and ABC1-C in yeast, reinforcing their secondary loss from insects. The secondary loss of conserved kinases within each lineage highlights how essential functions are dependent on the context of other genes and pathways in the organism.
Table 2. Kinase subfamilies shared by worm and human, but not fly.
| Group | Family | Subfamily | Worm genes | Human genes | Notes |
|---|---|---|---|---|---|
| AGC | SGK | 1 | 3 | Close relative to the AKT (PKB) family | |
| Atypical | ABC1 | ABC1-C | 1 | 1 | Other ABC1 subfamilies may compensate. |
| Atypical | MHCK | eEF2K | 1 | 1 | Eukaryotic elongation factor 2 kinase. |
| CAMK | CAMKL | MELK | 1 | 1 | MELK is an outgroup of MARK, which is present in fly. Splicing function? |
| CAMK | PSK | 1 | 2 | Human PSKH1 has a Golgi function. | |
| CAMK | PIM | 2 | 3 | Related to PASK, which is present in fly and absent from worm. | |
| TKL | MLK | HH498 | 1 | 1 | Human form is cardiac-specific, worm is neuronal-restricted. Divergent functions? |
| TK | Trk | 1 | 3 | Neurotrophin receptor. Fly has closely related Ror and Musk families. | |
| TK | Met | 2 | 2 | Worm has a clear Met homolog and a divergent family member. |
The C. elegans kinome is also marked by a dramatic expansion of a small number of kinase classes. C. elegans has almost twice as many kinases as Drosophila (438 vs. 241 genes), but virtually all the difference (195 of 197 genes) is accounted for by expansions of a small number of families and of worm-specific families.
Fifteen kinase subfamilies are nematode-specific, accounting for almost a quarter of the kinome (105 genes). They include 8 distinct subfamilies within a large expansion of CK1 group kinases, containing 78 kinases, and two FGFR-like receptor tyrosine kinase families. An additional 5 families are in the Other group, and have very little similarity to any non-worm kinases. In general, they are not well characterized.
Table 3. Worm-specific and worm-expanded kinase classes. Counts of genes in kinomes. C. briggsae data from unpublished analysis of genome-predicted peptides.
| Name/Classification | C. elegans | C. briggsae | Fly | Human |
|---|---|---|---|---|
| CK1/Dual | 3 | 3 | 0 | 0 |
| CK1/TTBKL | 31 | 22 | 0 | 0 |
| CK1/Worm6 | 28 | 19 | 0 | 0 |
| CK1/Worm7 | 2 | 1 | 0 | 0 |
| CK1/Worm8 | 3 | 1 | 0 | 0 |
| CK1/Worm9 | 2 | 0 | 0 | 0 |
| CK1/Worm10 | 2 | 2 | 0 | 0 |
| CK1/Worm11 | 1 | 2 | 0 | 0 |
| CK1/Unique | 6 | 3 | 0 | 0 |
| TK/Fer | 38 | 24 | 1 | 2 |
| RGC group | 27 | 20 | 6 | 5 |
| TK/KIN-16 | 16 | 6 | 0 | 0 |
| Other/Haspin | 13 | 1 | 1 | 1 |
| CMGC/GSK3 | 7 | 6 | 3 | 2 |
| CAMK/CAMKL/ CHK1 | 7 | 1 | 1 | 1 |
| Ste/Ste7 | 10 | 8 | 4 | 7 |
| CMGC/MAPK/Jnk | 5 | 3 | 1 | 3 |
| TK/KIN-9 | 5 | 5 | 0 | 0 |
| Other/Worm1 | 2 | 1 | 0 | 0 |
| Other/Worm2 | 3 | 2 | 0 | 0 |
| Other/Worm3 | 2 | 1 | 0 | 0 |
| Other/Worm4 | 1 | 1 | 0 | 0 |
| Other/Worm5 | 3 | 0 | 0 | 0 |
| Total | 217 | 132 | 17 | 21 |
These kinases may hold a key to several nematode-specific biological functions. In several cases, the expansions appear recent, as the members are closely related by sequence and chromosomal location, and several appear to have been generated since the C. elegans/C. briggsae split. Many may have reduced or no function: several have lost catalytic or other conserved residues and 20 of the 25 worm kinase pseudogenes are from these families, indicating a high rate of gene turnover. A similar expansion is seen in C. briggsae, and though it appears that this is more modest, some of this may be due to the preliminary nature of the annotation and kinase analysis of this genome.
Reproductive functions often drive rapid evolution, and there is some evidence implicating kinase expansions in nematode spermatogeneis. One CK1 gene (spe-6) and one Fer gene (spe-8) function in spermatogenesis, and half or more of these classes are selectively expressed in sperm by microarray analysis (Muhlrad and Ward, 2002; Reinke et al., 2000; P. Muhlrad and S. Ward, pers. commun.).
Receptor guanylate cyclases (RGC) have a catalytically inactive kinase domain, and have separately expanded in all three metazoans, but most dramatically in worm (Morton, 2004). Most are uncharacterized, but several are expressed in highly restricted sets of neurons and are implicated in chemosensation, and one (daf-11) is involved in dauer formation (Vowels and Thomas, 1992).
The KIN-9 (previously known as kin-6) and KIN-16 families encode receptor tyrosine kinases, whose kinase domains and overall structure most resemble the FGF receptor family. Some of the KIN-16 family have arrays of extracellular immunoglobulin repeats, and some have vestigial extracellular regions (Popovici et al., 1999), while KIN-6 members have diverse novel extracellular regions. KIN-16 includes the old-1 and old-2 genes thought to be involved in age and stress resistance (Rikke et al., 2000). Many of the KIN-16 genes are chromosomally clustered, and are poorly conserved in C. briggsae, indicating a recent origin (Figure 2A). The KIN-9 genes are not clustered and all have briggsae orthologs.
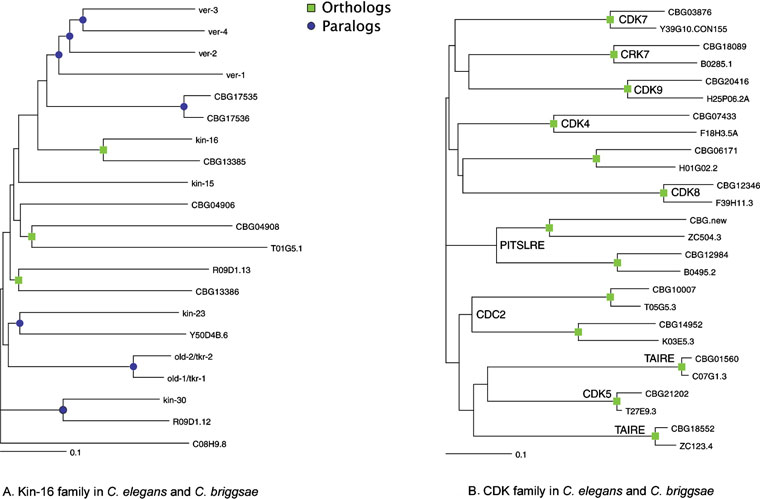
Figure 2. Orthology between C. elegans and C. briggsae kinases. Squares indicate likely orthologous pairs of kinases, and circles denote paralogous expansions. C. briggsae sequences are predictions (CBGnnnnn) from the genome project. (A) In the worm-specific KIN-16 family, new genes continue to arise in the elegans and briggsae lineages, as indicated by circles. (B) In the CDK family, all 13 members exist as orthologous pairs and subfamilies (labeled) are also conserved in Drosophila and human.
Of the 7 GSK3 members, 6 have clear briggsae homologs, but only one (gsk-3) has been characterized, and it acts in a defined Wnt signaling pathway (Schlesinger et al., 1999). The additional members may act in an expanded Wnt-like pathway, as worms have other duplicated pathway members including three members each of the beta-catenin and dishevelled families. Both CK1 and Fer kinases are implicated in mammalian Wnt signaling, and some of their worm expansions may also function in this pathway. The role of GSK3 in insulin signaling may also correlate with the expansion of insulin genes in nematodes.
The expansion of the Jnk stress-response MAPK family is partially paralleled by the expansion of the MAPKK (Ste7) family, which now includes 4 putative Jnk kinases (JNKKs).
A preliminary analysis of C. briggsae predicted proteins (release 25; Stein et al., 2003) indicates the presence of 341 kinase genes, using the C. elegans kinome as blast query set. An additional 30 or more kinases or kinase fragments were detected by direct search of the genome, but are still poorly predicted. The majority (320) of C. briggsae kinases appear orthologous to a single C. elegans kinase, by bidirectional blast searches. The main differences between the two kinomes are in the recently-expanded families, where the expansion appears to have continued since the elegans/briggsae split. Of 21 briggsae-unique kinases, 19 are from expanded families, and 98 of 117 elegans-unique kinases are from expanded families. More thorough sequence analysis will likely reveal more briggsae kinases and more ortholog pairs, but this data does strongly support both continued gene birth and death, and sequence diversification, within these expanded families. The difference between conserved and expanded families is shown in Figure 2A of the nematode-specific KIN-16 family, in which few pairs of obvious orthologs are seen between the two species. By contrast, the CDK family has 13 members in both species, all of which pair off in an orthologous fashion (Figure 2B).
Phosphatases remove phosphates from kinase substrates, both reversing kinase-based activation, and relieving kinase-mediated repressions. Phosphatases belong to several different families, including a number of distinct phosphatase domains: the PTP (protein tyrosine phosphatase), DSP (dual-specificity phosphatase which dephosphorylates both tyrosine and serine/threonine) and several families of STP, or serine-threonine phosphatases. While ‘phosphatome’ analysis lags behind that of the kinomes, an initial survey of C. elegans phosphatases identified 83 PTPs, 26 DSPs and 65 STPs (Plowman et al., 1999). The completion of two nematode genomes and multiple other eukaryotic genomes now opens the door for comparative analysis to identify additional C. elegans phosphatases and to compare their distribution with those of other organisms, and with the expansions of their cognate kinomes.
Manning, G., Plowman, G.D., Hunter, T., and Sudarsanam, S. (2002a). Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27, 514–520. Abstract Article
Manning, G., Whyte, D.B., Martinez, R., Hunter, T., and Sudarsanam, S. (2002b). The protein kinase complement of the human genome. Science 298, 1912–1934. Abstract Article
Morton, D.B. (2004). Invertebrates yield a plethora of atypical guanylyl cyclases. Mol. Neurobiol. 29, 97–116. Abstract
Muhlrad, P.J., and Ward, S. (2002). Spermiogenesis initiation in Caenorhabditis elegans involves a casein kinase 1 encoded by the spe-6 gene. Genetics 161, 143–1550. Abstract
Plowman, G.D., Sudarsanam, S., Bingham, J., Whyte, D., and Hunter, T. (1999). The protein kinases of Caenorhabditis elegans: a model for signal transduction in multicellular organisms. Proc. Natl. Acad. Sci. USA 96, 13603–13610. Abstract Article
Popovici, C., Roubin, R., Coulier, F., Pontarotti, P., and Birnbaum, D. (1999). The family ofCaenorhabditis elegans tyrosine kinase receptors: similarities and differences with mammalian receptors. Genome Res. 9, 1026–1039. Abstract
Reinke, V., Smith, H.E., Nance, J., Wang, J., Van Doren, C., Begley, R., Jones, S.J., Davis, E.B., Scherer, S., Ward, S., and Kim, S.K. (2000). A global profile of germline gene expression in C. elegans. Mol. Cell 6, 605–616. Abstract
Rikke, B.A., Murakami, S., and Johnson, T.E. (2000). Paralogy and orthology of tyrosine kinases that can extend the life span of Caenorhabditis elegans. Mol. Biol. Evol. 17, 671–683. Abstract
Schlesinger, A., Shelton, C.A., Maloof, J.N., Meneghini, M., and Bowerman, B. (1999). Wnt pathway components orient a mitotic spindle in the early Caenorhabditis elegans embryo without requiring gene transcription in the responding cell. Genes Dev. 13, 2028–2038. Abstract
Stein, L.D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M.R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. (2003). The genome sequence of Caenorhabditis briggsae: a platform for comparative genomics. PLoS Biol. 1, E45. Abstract Article
Vowels, J.J., and Thomas, J.H. (1992). Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics 130, 105–123. Abstract
Table 4. Classification of worm kinases
| Group | Family | Subfamily | # Kinases (domains) | Distribution | Names | Other Wormbook entries | Name/ function overview |
|---|---|---|---|---|---|---|---|
| AGC | 2 | Nematodes, Dictyostelium | F31E3.2, F28C10.3 | ||||
| AGC | AKT | 2 | All kinomes | akt-1, akt-2 | PI3K signaling | ||
| AGC | DMPK | GEK | 1 | All metazoans | K08B12.5 | Myotonic dystrophy protein kinase | |
| AGC | DMPK | ROCK | 1 | Metazoans, Dictyostelium | let-502 | Myotonic dystrophy protein kinase/Rho kinase | |
| AGC | GRK | BARK | 1 | All metazoans | grk-2 | Beta adrenergic receptor kinase | |
| AGC | GRK | GRK | 1 | All metazoans | grk-1 | G protein coupled kinase | |
| AGC | MAST | 1 | Metazoans, Dictyostelium | kin-4 | Microtubule associated serine/ threonine kinase | ||
| AGC | NDR | 2 | All kinomes | sax-1, T20F10.1 | |||
| AGC | PDK1 | 2 | All kinomes | pdk-1, W04B5.5 | PI3K signaling | ||
| AGC | PKA | 2 | All kinomes | kin-1, F47F2.1 | cAMP-dependent protein kinase | ||
| AGC | PKC | Alpha | 1 | All metazoans | pkc-2 | Protein kinase C isoforms | |
| AGC | PKC | Delta | 1 | All metazoans | tpa-1 | Protein kinase C isoforms | |
| AGC | PKC | Eta | 1 | All metazoans | kin-13 | Protein kinase C isoforms | |
| AGC | PKC | Iota | 1 | All metazoans | pkc-3 | Protein kinase C isoforms | |
| AGC | PKG | 2 | All metazoans | egl-4, C09G4.2 | cGMP-dependent protein kinase | ||
| AGC | PKN | 1 | All metazoans | F46F6.2 | Protein kinase N | ||
| AGC | RSK | MSK | 1 | All metazoans | C54G4.1 | Ribosomal S6 kinase | |
| AGC | RSK | p70 | 2 | Metazoans, fungi | Y43D4A.6, R04A9.5 | Ribosomal S6 kinase | |
| AGC | RSK | RSK | 1 | All metazoans | T01H8.1a | Ribosomal S6 kinase | |
| AGC | RSKL | 1 | All metazoans | F55C5.7 | Ribosomal S6 kinase-like | ||
| AGC | SGK | 1 | Nematodes, vertebrates, dictyostelium | W10G6.2 | Serum/glucocorticoid-regulated kinase | ||
| AGC | YANK | 1 | All metazoans | M03C11.1 | Uncharacterized | ||
| Atypical | A6 | 2 | All kinomes | unc-60, F38E9.5 | |||
| Atypical | ABC1 | ABC1-A | 1 | All kinomes | C35D10.4 | ||
| Atypical | ABC1 | ABC1-B | 1 | All kinomes | D2023.6 | ||
| Atypical | ABC1 | ABC1-C | 1 | All but insects | Y32H12A.7 | ||
| Atypical | Alpha | eEF2K | 1 | Nematodes, vertebrates, dictyostelium | efk-1 | Elongation factor 2 kinase | |
| Atypical | BRD | 3 | Metazoans, Dictyostelium | Y119C1B.8, F57C7.1b, F13C5.2 | Bromodo-main-containing kinase | ||
| Atypical | PDHK | 2 | Metazoans, fungi | ZK370.5, aSWK467 | Pyruvate dehydrogenase kinase | ||
| Atypical | PIKK | ATM | 1 | Metazoans, fungi | atm-1 | DNA damage response | |
| Atypical | PIKK | ATR | 1 | All kinomes | atl-1 | DNA damage response | |
| Atypical | PIKK | FRAP | 1 | All kinomes | B0261.2 | Metabolic regulation (aka TOR) | |
| Atypical | PIKK | SMG1 | 1 | Metazoans, Dictyostelium | smg-1 | mRNA surveillance | |
| Atypical | PIKK | TRRAP | 1 | All kinomes | C47D12.1 | ||
| Atypical | RIO | RIO1 | 1 | All kinomes | M01B12.5 | ||
| Atypical | RIO | RIO2 | 1 | All kinomes | Y105E8B.3 | ||
| Atypical | RIO | RIO3 | 1 | All metazoans | ZK632.3 | ||
| Atypical | TAF1 | 1 | All kinomes | taf-1 | Basal transcription: TFIID associated factor. | ||
| CAMK | CAMK1 | 1 | All kinomes | cmk-1 | Calmodulin-dependent protein kinase 1 | ||
| CAMK | CAMK2 | 1 | All metazoans | unc-43 | Calmodulin-dependent protein kinase 1 | ||
| CAMK | CAMKL | AMPK | 2 | All kinomes | aak-1, aak-2 | Metabolic regulation | |
| CAMK | CAMKL | BRSK | 1 | Metazoans, Dictyostelium | sad-1 | Neuronal vesicle release | |
| CAMK | CAMKL | CHK1 | 5 (7) | Metazoans, fungi | chk-1, Y43D4A.6, R02C2.1, R02C2.2, DC2.7 | Cell cycle checkpoint kinase 1 | |
| CAMK | CAMKL | LKB | 1 | Metazoans, Dictyostelium | par-4 | Activator of AMPK | |
| CAMK | CAMKL | MARK | 2 | All kinomes | par-1, F23C8.8 | Asymmetric cell division and axis formation in the embryo | Microtubule affinity regulating kinase |
| CAMK | CAMKL | MELK | 1 | Nematodes and vertebrates | W03G1.6 | Maternal embryonic leucine zipper kinase | |
| CAMK | CAMKL | NIM1 | 1 | All metazoans | F49C5.4 | ||
| CAMK | CAMKL | NuaK | 1 | All metazoans | B0496.3 | Uncharacterized | |
| CAMK | CAMKL | QIK | 1 | Metazoans, Dictyostelium | kin-29 | Qin induced kinase | |
| CAMK | CAMKL | SNRK | 1 | All metazoans | ZK524.4 | ||
| CAMK | CASK | 1 | All metazoans | lin-2 | |||
| CAMK | DAPK | 1 | All metazoans | K12C11.4 | Death-associated protein kinase | ||
| CAMK | DCAMKL | 2 | All metazoans | zyg-8, F32D8.1 | Doublecortin and CAMK-like | ||
| CAMK | MAPKAPK | MAPKAPK | 2 | All metazoans | K08F8.1, C44C8.6 | MAPK activated protein kinase | |
| CAMK | MAPKAPK | MNK | 1 | All metazoans | R166.5 | MAPK activated protein kinase | |
| CAMK | MLCK | 4 (5) | All metazoans | unc-22, C24G7.5, ZC373.4, F12F3.2 | Myosin light chain kinase | ||
| CAMK | PHK | 1 | All metazoans | Y50D7A.3 | Phosphorylase kinase | ||
| CAMK | PIM | 2 | Nematodes and vertebrates | prk-1, prk-2 | |||
| CAMK | PKD | 2 | All metazoans | T25E12.4, W09C5.5 | Protein kinase D | ||
| CAMK | PSK | 1 | Nematodes and vertebrates | R06A10.4 | Protein serine kinase | ||
| CAMK | RAD53 | 2 | Metazoans, fungi | chk-2, T08D2.7 | DNA damage checkpoint | ||
| CAMK | RSKb | MSKb | 0(1) | All metazoans | C54G4.1 | Second domain of RSK kinases | |
| CAMK | RSKb | RSKb | 0(1) | All metazoans | T01H8.1a | Second domain of RSK kinases | |
| CAMK | TSSK | 3 | All metazoans | B0511.4, W02B12.12, Y38H8A.4 | Testis-specific serine kinase | ||
| CK1 | Unique | 6 | Some metazoans | T15B12.2, ZK507.3, C25H3.1, F16B12.5, ZK507.1, K09E4.1 | |||
| CK1 | CK1 | CK1-A | 1 | All metazoans | C03C10.1 | Cell kinase 1/Casein kinase 1 | |
| CK1 | CK1 | CK1-D | 1 | All kinomes | F46F2.2 | Cell kinase 1/Casein kinase 1 | |
| CK1 | CK1 | CK1-G | 1 | Metazoans, fungi | Y106G6E.6 | Cell kinase 1/Casein kinase 1 | |
| CK1 | Dual | 3 (6) | Nematodes | F59A6.4, T05A7.6, H05L14.1 | Dual-domain CK1 kinase subfamily | ||
| CK1 | TTBK | 1 | Metazoans, Dictyostelium | R90.1 | Tau tubulin kinase | ||
| CK1 | TTBKL | 31 | Nematodes | M7.7, B0207.7, F35C11.3, Y71F9AL.2, C04G2.2, C45G9.1, F32B6.10, W01B6.2, C05C12.1, C49C8.1, Y73B6A.2, D2024.1, Y47G6A.13, F54H5.2, C56C10.6, C53A5.4, K06H7.8, D2045.5, W09C3.1, R10D12.10, T11F8.4, ZC581.2, T05C12.1, W06F12.3, C03C10.2, T19D12.5, ZK666.8, ZK354.6, F26A1.4, C14A4.13, W03G9.5 | Tau tubulin kinase-like | ||
| CK1 | VRK | 1 | All metazoans | F28B12.3 | Vaccinia-related kinase | ||
| CK1 | Worm10 | 2 | Nematodes | F26A1.3, F25F2.1 | Uncharacterized | ||
| CK1 | Worm11 | 1 | Nematodes | K11C4.1 | Uncharacterized | ||
| CK1 | Worm6 | 28 | Nematodes | Y38H8A.3, C39H7.1, ZK596.2, C50F4.10, C08F8.6, F36H12.8, R13H9.5, Y69F12A.1, B0218.5, F36H12.9, R13H9.6, T09B4.7, C09D4.3, C55B7.10, F41G3.5, F38E1.3, C27D8.1, Y39G8C.2, F53C3.1, F33D11.7, C34B2.3, C49C3.2, spe-6, C09B9.4, ZK354.2, Y65B4A.9, F59E12.3, C38C3.4 | Uncharacterized | ||
| CK1 | Worm7 | 2 | Nematodes | T01H10.4, ZC373.3 | Uncharacterized | ||
| CK1 | Worm8 | 3 | Nematodes | F22F1.2, F39F10.3, F39F10.2 | Uncharacterized | ||
| CK1 | Worm9 | 2 | Nematodes | K04C1.5, E02H4.6 | Uncharacterized | ||
| CMGC | 1 | Nematodes, fungi | F52B5.2 | ||||
| CMGC | CDK | 1 | Metazoans, fungi | H01G02.2 | Cyclin dependent kinase | ||
| CMGC | CDK | CDC2 | 2 | All kinomes | cdk-1, K03E5.3 | Cell cycle: cyclin dependent kinase | |
| CMGC | CDK | CDK4 | 1 | All metazoans | cdk-4 | Cell cycle: cyclin dependent kinase | |
| CMGC | CDK | CDK5 | 1 | All metazoans | cdk-5 | Cyclin dependent kinase | |
| CMGC | CDK | CDK7 | 1 | All kinomes | cdk-7 | Cell cycle: cyclin dependent kinase | |
| CMGC | CDK | CDK8 | 1 | All kinomes | F39H11.3 | Cell cycle: cyclin dependent kinase | |
| CMGC | CDK | CDK9 | 1 | All metazoans | H25P06.2 | Cell cycle: cyclin dependent kinase | |
| CMGC | CDK | CRK7 | 1 | All kinomes | B0285.1 | Cell cycle: cyclin dependent kinase | |
| CMGC | CDK | PITSLRE | 2 | Metazoans, Dictyostelium | ZC504.3, B0495.2 | Cell cycle: cyclin dependent kinase | |
| CMGC | CDK | TAIRE | 2 | All metazoans | pct-1, ZC123.4 | Cell cycle: cyclin dependent kinase | |
| CMGC | CDKL | 1 | All metazoans | Y42A5A.4 | Cyclin dependent kinase-like | ||
| CMGC | CLK | 3 | All kinomes | C16A11.3, Y111B2A.1, E02H4.3 | CDC-like kinase, involved in splicing | ||
| CMGC | Dyrk | Dyrk1 | 1 | Metazoans, Dictyostelium | mbk-1 | ||
| CMGC | Dyrk | Dyrk2 | 3 | Metazoans, Dictyostelium | mbk-2, C36B7.1, C36B7.2 | ||
| CMGC | Dyrk | HIPK | 1 | All metazoans | hpk-1 | Hypoxia-inducible protein kinase | |
| CMGC | Dyrk | PRP4 | 1 | Metazoans, Dictyostelium | F22D6.5 | mRNA processing | |
| CMGC | GSK | 7 | All kinomes | gsk-3, R03D7.5, Y106G6D.4, C44H4.6, Y106G6E.1, C36B1.10, F21F3.2 | Glycogen synthase kinase 3 | ||
| CMGC | MAPK | 3 | Some metazoans | W06B3.2, F09C12.2, C04G6.1 | |||
| CMGC | MAPK | Erk | 1 | All kinomes | mpk-1 | RTKRas/ MAP kinase signaling | Growth factor response MAPK |
| CMGC | MAPK | Erk7 | 1 | Metazoans, Dictyostelium | C05D10.2 | Variant MAPK | |
| CMGC | MAPK | Jnk | 5 | All metazoans | jnk-1, T07A9.3, Y51B9A.9, ZC416.4, C49C3.10 | Signaling in the immune response | Stress-response MAPK |
| CMGC | MAPK | nmo | 1 | All metazoans | lit-1 | Variant MAPK | |
| CMGC | MAPK | p38 | 3 | Metazoans, fungi | pmk-1, pmk-2, pmk-3 | Stress-response MAPK | |
| CMGC | RCK | 1 | All kinomes | M04C9.5 | |||
| CMGC | SRPK | 1 | All kinomes | spk-1 | mRNA splicing | ||
| Other | Unique | 10 | All kinomes | flr-4, zyg-1, Y106G6A.1, D2045.7, K09A9.1, Y53F4B.1, K11H12.9, K02E10.7, F37E3.3, C29H12.5 | |||
| Other | Aur | 2 | All kinomes | air-1, air-2 | Cell cycle/chromosomal stability | ||
| Other | Bub | 1 | All kinomes | bub-1 | Cell cycle | ||
| Other | Bud32 | 1 | All kinomes | F52C12.2 | Essential homolog of yeast Bud32. | ||
| Other | CAMKK | Meta | 1 | Metazoans, Dictyostelium | ckk-1 | CAMK kinase | |
| Other | CDC7 | 1 | All kinomes | C34G6.5 | Variant cyclin-dependent kinase | ||
| Other | CK2 | 1 | All kinomes | B0205.7 | Cell kinase 2/Casein kinase 2 | ||
| Other | Haspin | 13 | All kinomes | Y18H1A.10, K08B4.5, C50H2.7, T05E8.2, ZK177.2, H12I13.1, Y48B6A.10, C01H6.9, F22H10.5, C04G2.10, Y40A1A.1, Y73B6A.1, aSWK457 | Nuclear kinase; meiosis? | ||
| Other | IRE | 1 | All kinomes | ire-1 | Unfolded protein response | ||
| Other | Nak | 2 | All kinomes | sel-5, F46G11.3 | |||
| Other | Nek | 4 | All kinomes | F19H6.1, ZC581.1, Y39G10AR.3, pqn-25, | Cell cycle (NIMA-related kinase) | ||
| Other | NKF1 | 1 | All metazoans | C01C4.3 | Uncharacterized | ||
| Other | NKF2 | 1 | All metazoans | EEED8.9 | Uncharacterized | ||
| Other | NRBP | 1 | All metazoans | K10D3.5 | Nuclear receptor binding protein | ||
| Other | PEK | GCN2 | 1 | All kinomes | Y81G3A.3 | ||
| Other | PEK | PEK | 2 | Metazoans, Dictyostelium | pek-1, Y38E10A.8 | ||
| Other | PLK | 3 | All kinomes | plk-1, plk-2, F55G1.8 | Cell cycle: polo-like kinase | ||
| Other | SCY1 | 2 | All kinomes | W07G4.3, ZC581.9 | |||
| Other | TBCK | 1 | Metazoans, Dictyostelium | C33F10.2 | TBC domain kinase | ||
| Other | TLK | 1 | All metazoans | C07A9.3 | Tousled-like kinase | ||
| Other | ULK | 2 | All kinomes | unc-51, T07F12.4 | Homolog of Drosophila Fused | ||
| Other | VPS15 | 1 | All kinomes | ZK930.1 | |||
| Other | WEE | 2 | All kinomes | wee-1.1, wee-1.3 | Cell cycle | ||
| Other | Wnk | 1 | All metazoans | C46C2.1 | With no lysine[K] | ||
| Other | Worm1 | 2 | Nematodes | mes-1, B0198.3 | Uncharacterized | ||
| Other | Worm2 | 3 | Nematodes | K09C6.8, T10B5.2, K09C6.7 | Uncharacterized | ||
| Other | Worm3 | 2 | Nematodes | R107.4, Y39G8B.5 | Uncharacterized | ||
| Other | Worm4 | 1 | Nematodes | C28A5.6 | Uncharacterized | ||
| Other | Worm5 | 3 | Nematodes | C44C10.7, F16B12.7, K08H2.5 | Uncharacterized | ||
| RGC | RGC | 27 | All metazoans | gcy-13, gcy-14, odr-1, gcy-19, gcy-15, gcy-9, daf-11, T01A4.1b, gcy-4, gcy-17, gcy-3, gcy-7, C04H5.4a, gcy-11, gcy-18, gcy-25, gcy-21, gcy-27, gcy-23, gcy-1, gcy-20, gcy-5, gcy-6, gcy-8, gcy-22, gcy-2, gcy-12 | Receptor guanylate cyclase | ||
| STE | Unique | 1 | Metazoans, Dictyostelium | F18F11.4 | |||
| STE | Ste11 | 2 | All kinomes | nsy-1, B0414.7 | MAP kinase kinase kinase | ||
| STE | Ste20 | FRAY | 1 | Metazoans, Dictyostelium | Y59A8B.23 | MAP4 kinase and related | |
| STE | Ste20 | KHS | 1 | All metazoans | ZC404.9 | Subfamily of MAP4K kinases | |
| STE | Ste20 | MSN | 1 | All metazoans | ZC504.4 | Subfamily of MAP4K kinases | |
| STE | Ste20 | MST | 1 | All kinomes | F14H12.4 | Subfamily of MAP4K kinases | |
| STE | Ste20 | PAKA | 2 | All kinomes | C09B8.7, Y38F1A.1 | Couples small GTPases to cytoskeleton | |
| STE | Ste20 | PAKB | 1 | All metazoans | C45B11.1 | Couples small GTPases to cytoskeleton | |
| STE | Ste20 | SLK | 1 | All metazoans | C04A11.3 | Subfamily of MAP4K kinases | |
| STE | Ste20 | STLK | 2 | All metazoans | C03B1.5, Y52D3.1 | Subfamily of MAP4K kinases | |
| STE | Ste20 | TAO | 1 | All metazoans | T17E9.1 | Subfamily of MAP4K kinases | |
| STE | Ste20 | YSK | 1 | Metazoans, fungi | T19A5.2 | Subfamily of MAP4K kinases | |
| STE | Ste7 | 10 | All kinomes | jkk-1, mek-1, mek-2, sek-1, mkk-4, ZC449.6, VZC374L.1, E02D9.1a, F35C8.2, F35C8.1 | RTKRas/ MAP kinase signaling | MAP kinase kinase | |
| TK | Unique | 7 | All metazoans | C16D9.2, Y38H6C.20, F40A3.5, F59F5.3, R151.4, C34F11.5, T22B11.3 | |||
| TK | Abl | 1 | All metazoans | abl-1 | Abelseon leukemia virus homolog | ||
| TK | Ack | 2 | All metazoans | ark-1, kin-25 | |||
| TK | Csk | 1 | All metazoans | Y48G1C.2 | |||
| TK | Fer | 38 | All metazoans | frk-1, spe-8, kin-5, kin-14, kin-21, kin-24, kin-26, kin-28, aSWK454, Y43C5B.2, Y116A8C.24, K09B11.5, Y52D5A.2, Y69E1A.3, T25B9.5, M176.9, ZC581.7, F59A3.8, R05H5.4, W01B6.5, T08G5.2, F57B9.8, T21G5.1, C25A8.5, T06C10.3, F22B3.8, F01D4.3, W03F8.2, ZK622.1, R11E3.1, T25B9.4, ZK593.9, F26E4.5, W03A5.1, C35E7.10, C55C3.4, C18H7.4, F23C8.7 | |||
| TK | Src | 3 | All metazoans | src-1, src-2, Y47G6A.5a | Non-receptor tyrosine kinase | ||
| TKL | IRAK | 1 | All metazoans | K09B11.1 | IL-1 receptor associated kinase | ||
| TKL | LRRK | 1 | Metazoans, Dictyostelium | T27C10.5 | Leucine rich repeat kinase | ||
| TKL | MLK | HH498 | 1 | Nematodes, vertebrates, Dictyostelium | C24A1.3 | Uncharacterized | |
| TKL | MLK | ILK | 1 | All metazoans | C29F9.7 | Integrin linked kinase | |
| TKL | MLK | LZK | 1 | All metazoans | F33E2.2 | ||
| TKL | MLK | MLK | 2 | Metazoans, Dictyostelium | Y58A7A.f, R13F6.7 | Mixed lineage kinase | |
| TKL | MLK | TAK1 | 2 | All metazoans | F52F12.3, Y105C5.y | ||
| TKL | RAF | 3 | All metazoans | raf-1, pex-1, ksr-1 | RTKRas/ MAP kinase signaling | Ras-MAPK signaling | |
| TKL | STKR | Type1 | 2 | All metazoans | sma-6, daf-1 | TGF-β signaling | TGFb and related receptors |
| TKL | STKR | Type2 | 1 | All metazoans | daf-4 | TGF-β signaling | TGFb and related receptors |
| TK-RTK | Alk | 1 | All metazoans | scd-2 | |||
| TK-RTK | DDR | 2 | All metazoans | F11D5.3, C25F6.4 | Discoidin domain receptor. | ||
| TK-RTK | EGFR | 1 | All metazoans | let-23 | RTKRas/ MAP kinase signaling | Growth factor (EGF) receptor | |
| TK-RTK | Eph | 1 | All metazoans | vab-1 | Ephrin receptor | ||
| TK-RTK | Fak | 1 | All metazoans | kin-32 | Focal adhesion kinase | ||
| TK-RTK | FGFR | 1 | All metazoans | egl-15 | RTKRas/ MAP kinase signaling | Growth factor FGF) receptor | |
| TK-RTK | InsR | 1 | All metazoans | daf-2 | Insulin receptor | ||
| TK-RTK | KIN-16 | 15 (16) | Nematodes | old-1, old-2, ver-1, R09D1.13, kin-23, T17A3.8, T01G5.1, C08H9.8, F59F3.1, M176.7, F59F3.5, M176.6, kin-30, R09D1.12, Y50D4B.6 | Uncharacterized | ||
| TK-RTK | KIN-9 | 5 | Nematodes | F09G2.1, F09A5.2, C24G6.2, F08F1.1, B0252.1 | Uncharacterized | ||
| TK-RTK | Met | 2 | Nematodes and vertebrates | F11E6.8, T14E8.1 | Growth factor (HGF/SF) receptor | ||
| TK-RTK | Ror | 1 | All metazoans | cam-1 | Receptor tyrosine kinase | ||
| TK-RTK | Ryk | 1 | All metazoans | C16B8.1 | |||
| TK-RTK | Trk | 1 | Nematodes and vertebrates | D1073.1 | Neurotropin receptor | ||
| TOTAL | 438(447) |
*Edited by Iva Greenwald. Last revised December 10, 2005. Published December 13, 2005. This chapter should be cited as: Manning, G. Genomic overview of Protein Kinases (December 13, 2005), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.60.1, http://www.wormbook.org.
§To whom correspondence should be addressed. E-mail: manning@salk.edu
 All journal content, except where otherwise noted, is licensed under a Creative Commons Attribution License.
All journal content, except where otherwise noted, is licensed under a Creative Commons Attribution License.