 Embryo series courtesy of Einhard Schierenberg
Embryo series courtesy of Einhard SchierenbergTable of Contents
Abstract
Sexual identity is one of a few basic parameters that specify how development should proceed. Although sex determination has profound effects on many tissues, its most ancient and fundamental role is ensuring that some germ cells become sperm, and others become oocytes or eggs. Spermatocytes and oocytes are usually produced in male and female animals, respectively, but C. elegans is uniquely suitable for studying the control of these cell fates because both types of cells are made from a common pool of progenitors in XX hermaphrodites. Extensive genetic and molecular studies have shown that the sexual fate of germ cells in C. elegans is controlled by the same genes that regulate sexual identity in other parts of the animal. However, this regulatory pathway has additional features that are unique to the germ line. First, several genes, like the three fogs, act only in germ cells. Second, the three fem genes act in concert with targets of tra-1 to control germ cell fates, but do not act this way in the soma. Third, translational repression of tra-2 is essential for hermaphrodite spermatogenesis. Fourth, translational repression of fem-3 is needed for oogenesis. In this review, we present genetic and molecular models for how these processes work, and summarize the evidence upon which they are built.
In C. elegans there are two sexes. Animals with two X chromosomes (XX) and two sets of autosomes have a female soma and a hermaphrodite germ line, and so are called hermaphrodites. Animals with a single X chromosome (XO) and two sets of autosomes have a male soma and germline (see Somatic sex determination). Hermaphrodite self-fertility is achieved by a transient period of spermatogenesis in the L3 stage followed by a switch to oogenesis in the L4. Males begin spermatogenesis in the L3/L4 stage and this continues through adulthood. Although one might expect that the regulation of sexual identity in germ cells would be just like female/male choices in the soma, studies in both worms and flies show that it is more complex. Several features that make the germ line unique might contribute to this complexity. For example, the germ line is the most ancient tissue in animals, and the only sexually dimorphic tissue common to all species. Thus, it has had a long evolutionary history and many opportunities to accumulate new regulatory circuits. In addition, oocytes or eggs need to be packed with regulatory molecules that will direct early embryonic development, including sex determination, and some of these molecules must be repressed so that they don’t affect the germ line itself. Finally, in species like C. elegans, where the germline of a single individual can produce both male and female germ cells, additional layers of regulation are required to ensure plasticity in sexual fate.
The core pathway that regulates somatic sexual fate forms a central part of the pathway that controls germline sexual fate (Figure 1; Goodwin and Ellis; 2002; see Somatic sex determination and X-chromosome dosage compensation). It is composed of seven genes that are each necessary for sexual identity in germ cells and somatic cells. These genes function in a negative regulatory hierarchy to control TRA-1 activity (Hodgkin, 1987, see Somatic sex determination). However, TRA-1 is not the terminal regulator of sexual fate in the germ line, and germline specific genes and regulatory interactions are superimposed on this core pathway to allow hermaphrodite development. Below, we briefly describe the core pathway and then the additional genes and layers of regulation that act in germline sex determination.
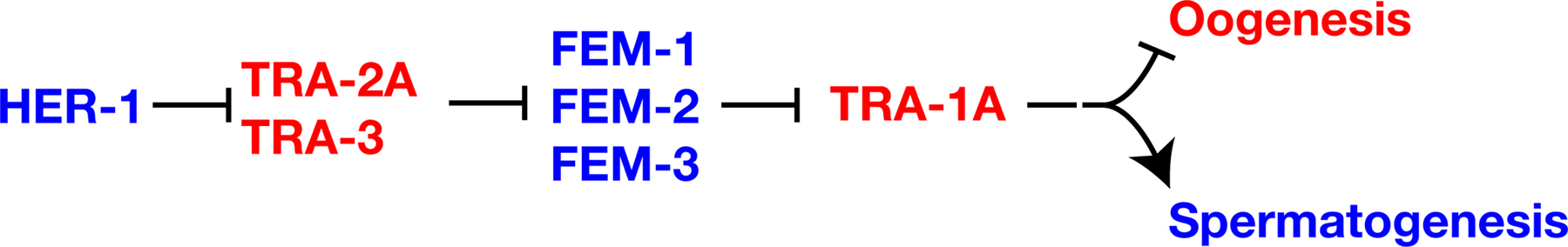 |
Figure 1. The core sex determination pathway. Proteins are shown in capital letters, and are indicated when the regulatory interactions are known to take place at this level; in all other cases, gene names are used and are shown in italics. Positive interactions are indicated with an arrow, and negative ones by a “--|”. Genes that promote spermatogenesis are blue, and those that promote oogenesis are red. Not shown are sex-determination and dosage compensation genes that act upstream of HER-1, which also form part of the “global” sex determination pathway (see Somatic sex determination and X-chromosome dosage compensation).
The her-1 gene acts in XO animals to promote male somatic development and continuous spermatogenesis (Hodgkin, 1980). This her-1 activity is restricted to XO animals by the SDC (sex determination and dosage compensation) genes, which repress her-1 transcription in XX animals (Chu et al., 2002; for details see Somatic sex determination and X-chromosome dosage compensation). HER-1 plays a pivotal role in coordinating sexual fate throughout the animal, as several lines of evidence indicate that HER-1 transmits a masculinizing signal between cells. This evidence includes sequence analyses indicating that HER-1 is a small, secreted protein (Perry et al., 1993), genetic mosaic analyses indicating that HER-1 acts non-autonomously (Hunter and Wood, 1992), and ectopic expression of HER-1 in the muscle of XX animals, which masculinizes somatic tissues and causes germ cells to become sperm (Perry et al., 1993). Based on mosaic analysis, HER-1 activity in the intestine and somatic gonad exerts the greatest influence on germ cell fates (Hunter and Wood, 1992).
HER-1 promotes spermatogenesis by binding to and inactivating the transmembrane protein TRA-2 (Kuwabara and Kimble, 1995; Kuwabara et al., 1992; Okkema and Kimble, 1991). Likely HER-1 sites required for this interaction have been identified by X-ray analysis of HER-1 crystals (Hamaoka et al., 2004), and the TRA-2 receptor site by genetic analysis (Kuwabara, 1996). TRA-2, in turn, inactivates the FEM proteins (Hodgkin, 1986). The cytoplasmic domain of TRA-2 can bind FEM-3 (Mehra et al., 1999). Thus, a simple model is that HER-1 binding to the extracellular domain of TRA-2 disrupts the ability of the intracellular domain of TRA-2 to bind and inactivate FEM-3. FEM-1 contains ankyrin repeats, FEM-2 is a PP2C phosphatase, and FEM-3 is novel (Ahringer et al., 1992; Pilgrim et al., 1995; Spence et al., 1990). TRA-1 is a transcription factor related to the Drosophila Ci and vertebrate GLI proteins (Zarkower and Hodgkin, 1992). The FEM proteins inactivate TRA-1 by a mechanism that is not yet understood.
In Drosophila, germ cell sexual fate is also controlled by somatic signals, and these signals interact with processes intrinsic to the germ line (reviewed by Oliver, 2002). In flies, additional genes act only in the germ line to control sexual fate, but the genetic pathways involved have been hard to unravel. By contrast, several well-characterized regulatory circuits have been identified in the nematode germ line; in general, these circuits modulate the activity of sex-determination genes to allow for hermaphrodite spermatogenesis.
In both males and hermaphrodites, sperm production requires fog-1, fog-3 and the three fem genes (Barton and Kimble, 1990; Doniach and Hodgkin, 1984; Ellis and Kimble, 1995; Hodgkin, 1986). Mutations in any of these genes cause all germ cells to differentiate as oocytes.
Two facts imply that fog-1 and fog-3 control sexual fate in germ cells and probably act at the end of the sex-determination pathway. First, mutations in fog-1 or fog-3 cause germ cells to differentiate as oocytes rather than sperm, but have no effect on other parts of the body. Second, genetic analyses indicate that mutations in fog-1 and fog-3 act downstream of mutations in all other genes in the pathway (Figure 2). Thus, these two genes might be directly responsible for initiating spermatogenesis.
The fog-1 gene produces two major transcripts (Jin et al., 2001; Luitjens et al., 2000). The large transcript is necessary for fog-1 function and is sufficient to rescue fog-1(q253) mutants, but the small one has no detectable activity (Jin et al., 2001a; Jin et al., 2001b). Other sex-determination genes, like her-1, tra-1 and tra-2, also make minor transcripts of unknown function. Here we discuss only the essential products of each gene.
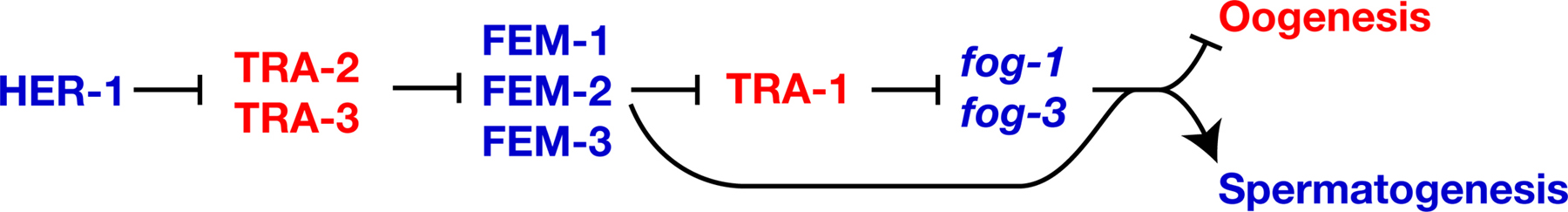 |
Figure 2. FOG-1, FOG-3 and the FEM proteins promote spermatogenesis. Proteins are shown in uppercase, and genes in lowercase italics, to indicate that TRA-1 controls transcription of the fog-1 and fog-3 genes. Positive interactions are indicated with an arrow, and negative ones by a “--|”. Genes that promote spermatogenesis are blue, and those that promote oogenesis are red.
Sequence analysis reveals that FOG-1 is a Cytoplasmic Polyadenylation Element Binding (CPEB) protein. All CPEB proteins contain two RNA Recognition Motifs (RRMs) and a C-H domain that binds zinc and interacts with RNA (reviewed by Mendez and Richter, 2001). Because other CPEB proteins regulate the translation of specific messages, FOG-1 probably regulates the translation of mRNAs that control sexual fate in the germ line. Interestingly, FOG-1 can bind the fog-1 message itself (Jin et al., 2001a), so it might be auto-regulatory. Other targets of FOG-1 have not yet been identified.
Although three other CPEB proteins are also expressed in the germ line (Luitjens et al., 2000), only FOG-1 regulates sexual fate. How is this specificity achieved? Analyses of FOG-1 homologs from other Caenorhabditis species show that FOG-1 has a conserved insertion in the C-H domain that is not found in other CPEB proteins (Cho et al., 2004); this domain might specify FOG-1’s unique function. One possibility is that FOG-1 binds specific elements found only in mRNAs that act in sex determination. Alternatively, FOG-1 might interact with other proteins that confer the ability to regulate sexual fate.
FOG-3 is a member of the Tob family of proteins (Chen et al., 2000). These proteins are found in all animals, and share a common amino-terminal domain of 115 amino acids. This family has been implicated in the control of cell division and differentiation, but so far little is know about their biochemical functions. Tob itself has both nuclear import and export signals in the conserved amino terminus (Kawamura-Tsuzuku et al., 2004; Maekawa et al., 2004), and associates with several Smad transcription factors during osteogenesis (Yoshida et al., 2000), so perhaps FOG-3 also regulates transcription.
TRA-1 appears to control germ cell fate by regulating the transcription of fog-1 and fog-3. Each gene has three to six potential TRA-1 binding sites in its promoter; more than are found in any other gene in the genome (Chen and Ellis, 2000; Jin et al., 2001). Furthermore, the fog-3 promoter can bind TRA-1 in gel shift assays, and this binding requires intact TRA-1 sites (Chen and Ellis, 2000). However, analyses of promoter mutations show that some of the TRA-1 binding sites mediate activation of fog-3 transgenes, whereas studies of fog-3 expression imply that at least one site mediates repression. Furthermore, tra-1(null) mutants usually make sperm early in life and oocytes later on (Hodgkin, 1987; Schedl et al., 1989). One explanation for these complex observations is that TRA-1 acts on the fog-3 promoter to repress spermatogenesis in some circumstances, and to activate it in others.
Just as in the soma, the fem genes regulate the activity of tra-1 in germ cells. For example, quantitative RT-PCR studies of fem; tra-1 double mutants show that the FEM proteins act through TRA-1 to control the level of fog-3 transcripts (Chen and Ellis, 2000). However, these double mutants make oocytes instead of sperm, which shows that the FEM proteins also act downstream of TRA-1 (Figure 2, Hodgkin, 1986). Perhaps the FEM proteins directly activate FOG-1 or FOG-3, or regulate an unknown target.
How do hermaphrodites undergo a brief period of spermatogenesis, given that XX animals do not express the HER-1 masculinizing non-autonomous signal? Genetic screens have revealed three regulatory modules that play a central role in this process.
Gain-of-function mutations in tra-2 cause XX animals to make only oocytes, but have little effect on males (Doniach, 1986). Sequence analysis showed that these tra-2(gf) mutations disrupt sequences in the 3’ UTR known as TGEs (Goodwin et al., 1993). In wild-type animals, these elements bind a protein complex that includes GLD-1 (Goodwin et al., 1993; Jan et al., 1999). Three observations imply that GLD-1 represses translation of tra-2 (Figure 3). First, GLD-1 is required for spermatogenesis in hermaphrodites but not in males (Francis et al., 1995; Francis et al.,1995). Second, somatically expressed GLD-1 can repress translation of somatic transgene RNAs that contain TGEs (Jan et al., 1999). Third, TRA-2 levels are increased in gld-1 null mutant germ lines, based on in situ antibody staining (Jan et al., 1999).
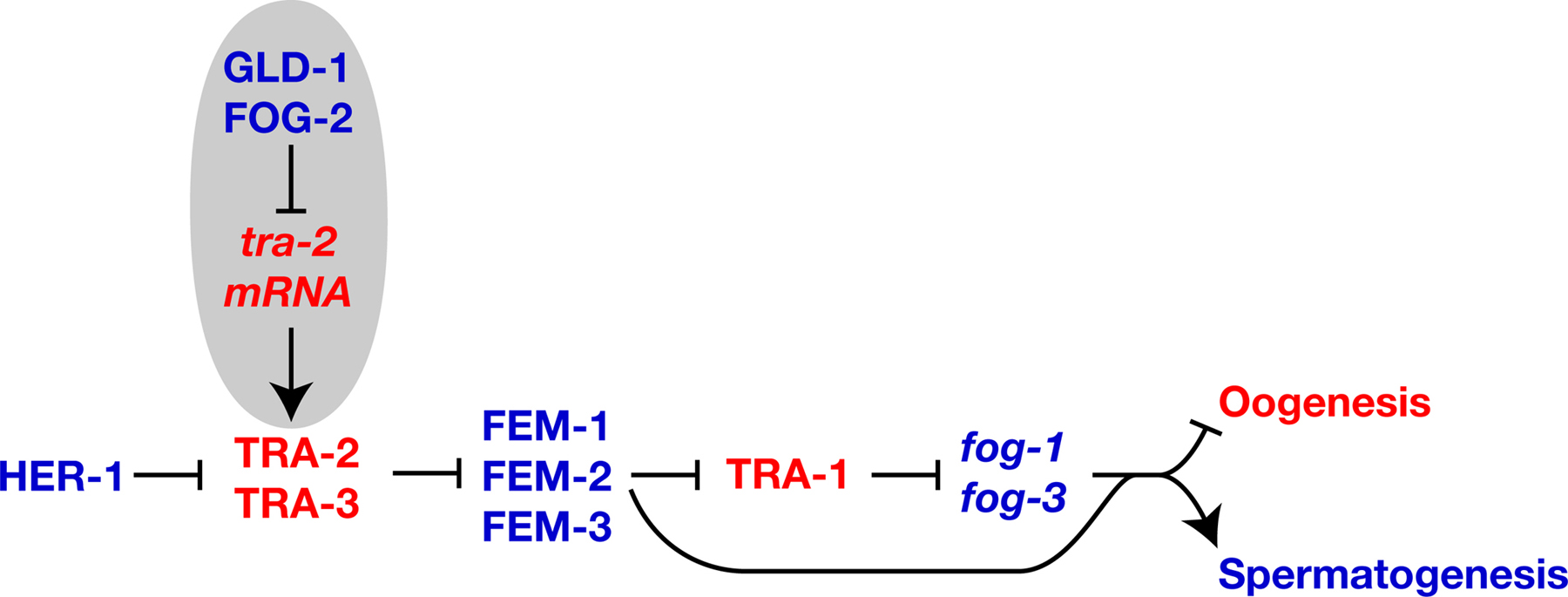 |
Figure 3. Regulatory modules that promote spermatogenesis. Proteins are shown in uppercase, and genes or mRNAs in lowercase italics. Positive interactions are indicated with an arrow, and negative ones by a “--|”. Genes that promote spermatogenesis are blue, and those that promote oogenesis are red. Regulatory circuits that promote hermaphrodite spermatogenesis are highlighted in gray.
GLD-1 is a member of the STAR family of proteins, which contain a maxi-KH RNA binding domain and two other conserved domains, called QUA1 and QUA2 (Jones et al., 1996). In vitro studies indicate that GLD-1 acts as a dimer (Ryder et al., 2004). Because GLD-1 has pleiotropic roles in germline development (Francis et al., 1995; Francis et al., 1995) it must regulate other messages in addition to tra-2. Some of these mRNA targets have been identified (Lee and Schedl, 2001; 2004; Marin and Evans, 2003; Mootz et al., 2004). Recently, in vitro biochemical studies implicated a hexanucleotide sequence as part of the GLD-1 binding site, at least for some RNA targets (Ryder et al., 2004).
fog-2 is required specifically for hermaphrodite spermatogenesis (Schedl and Kimble, 1988). FOG-2 binds GLD-1 in the yeast two-hybrid assay and in vitro, which implies that they interact to regulate tra-2 (Figure 3; Clifford et al., 2000). Genetic data support this model, since epistasis analyses place fog-2 upstream of tra-2, and loss of fog-2 activity causes the same phenotype as tra-2(gf) mutations (Schedl and Kimble, 1988). The C-terminus of FOG-2, which mediates its interaction with GLD-1, has been evolving rapidly (Nayak et al., 2004). The N-terminus of FOG-2 contains an F-box (Clifford et al., 2000) and binds the Skp1-related protein SKR-1 (Nayak et al., 2004); in other proteins, this would imply an interaction with the SCF ubiquitin ligase machinery. However, FOG-2 cannot target GLD-1 for degradation, since both promote spermatogenesis. Perhaps it recruits other proteins to the tra-2 3’UTR to block translation.
Some data suggest that laf-1 also regulates the translation of tra-2 messages (Goodwin et al., 1997). However, definitive tests of epistasis have not been possible, because laf-1 alleles cause recessive lethality as well as dominant feminization.
Several missense mutations in the intracellular domain of TRA-2 prevent spermatogenesis in hermaphrodites but not in males (Doniach, 1986; Kuwabara et al., 1998). Although this feminizing effect is dominant, these mutations also cause recessive masculinization in the soma, so they were named mixomorphic (mx) alleles. Surprisingly, these alleles alter a region of TRA-2 that normally binds TRA-1, and disrupt this interaction (Lum et al., 2000; Wang and Kimble, 2001). What remains unclear is the purpose of this TRA-2/TRA-1 interaction. One possibility is that the TRA-2/TRA-1 complex directly promotes oogenesis and other female fates; this model is consistent with the fact that tra-2(mx) alleles behave as if they cause increased wild-type activity (Doniach, 1986; Schedl and Kimble, 1988). Alternatively, the TRA-2/TRA-1 complex might promote spermatogenesis, as implied by the fact that blocking this interaction leads to oogenesis.
Since TRA-2 is a membrane protein, and TRA-1 is a transcription factor, how do they find each other? Lum et al. (2000) showed that the intracellular portion of TRA-2 is imported to the nucleus. Since tra-2 requires tra-3 to promote female development, and tra-3 encodes a calpain protease (Barnes and Hodgkin, 1996), TRA-3 might cleave TRA-2 to release this intracellular fragment. This interaction has been confirmed in vitro (Sokol and Kuwabara, 2000). Although germ cells produce a small transcript, tra-2B, that encodes a similar intracellular fragment, this transcript is associated with oogenesis rather than spermatogenesis (Kuwabara et al., 1998), and is unlikely to act in this regulatory process.
Since TRA-1 is a transcription factor, it needs to be in the nucleus to control cell fates. Surprisingly, TRA-1 binds sequences in the 3’-UTR of tra-2 messages (Graves et al., 1999). Furthermore, a mutation in tra-2 that blocks this interaction increases the levels of nuclear TRA-1 (Segal et al., 2001). This mutation also promotes oogenesis, though this effect is not unexpected since it deletes both TGEs, which are needed for translational repression of tra-2. Taken together, these results suggest that tra-2 messages might negatively regulate TRA-1 activity. The export of tra-2 messages requires CRM-1, as well as a group of genes that prevent export by the alternative NXF-1 pathway (Kuersten et al., 2004). Perhaps these complex interactions between tra-2 and TRA-1 help modulate TRA-1 activity during hermaphrodite development.
Hermaphrodites are only fertile if they eventually stop making sperm and begin to produce oocytes. By the mid-L4 stage, all germ cells that will form sperm are in pachytene (or later stages) and about the same number of cells have begun oogenesis, based on expression of the yolk receptor transcript (Figure 4). Thus, the decision to switch from spermatogenesis to oogenesis must have occurred at some earlier, yet to be determined, point. Three lines of evidence suggest that a crucial factor for determining how long hermaphrodites make sperm is the ratio of TRA-2 activity to FEM-3 activity. First, the tra-2 gain-of-function mutations described above increase the production of TRA-2, which causes XX animals to make oocytes instead of sperm (Doniach, 1986). Second, fem-3 gain-of-function mutations increase the production of FEM-3, which causes XX animals to make sperm instead of oocytes (below, Barton et al., 1987). Third, these tra-2(gf) and fem-3(gf) mutations can mutually suppress each other, restoring production of both sperm and oocytes (Schedl and Kimble, 1988). Because germ cell fates are so sensitive to the ratio of TRA-2 to FEM-3 activity, it is not surprising that additional regulatory modules control the production of FEM-3 protein from fem-3 messenger RNAs.
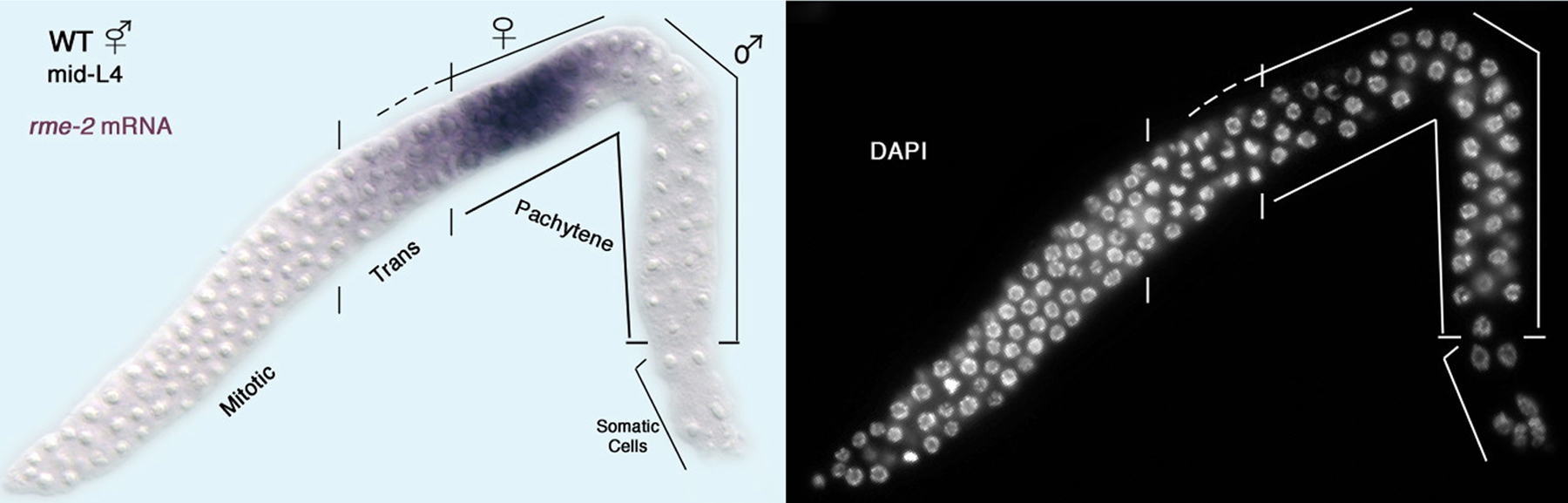 |
Figure 4. An early point following the switch from hermaphrodite spermatogenesis to oogenesis. In situ hybridization of the rme-2 yolk receptor mRNA to a dissected L4 hermaphrodite gonad: left, DIC image with rme-2 staining in purple; right, fluorescence microscopy image of DAPI staining to visualize chromosome morphology; distal tip on the lower left. Shown are the meiotic prophase stages leptotene and zygotene (indicated ‘trans’ for transition zone) as well as pachytene. Germ cells in the proximal end of the gonad are undergoing spermatogenesis and do not express the rme-2 yolk receptor mRNA. Germ cells located just distally, from the transition zone to the loop (bend in gonad) are undergoing oogenesis and express rme-2 mRNA (M. H. Lee and T. Schedl, unpublished results). Thus, at this time point, there are both male and female pachytene stage germ cells.
Hermaphrodites cannot switch from the production of sperm to that of oocytes unless fem-3 activity is repressed by a “Point Mutation Element,” or PME, located in the 3’UTR (Ahringer and Kimble, 1991; Ahringer et al., 1992; Rosenquist and Kimble, 1988). Gain-of-function mutations in fem-3 alter this PME, resulting in increased fem-3 activity and constitutive spermatogenesis. Since these mutations don’t change the level of fem-3 transcripts, but appear to increase the length of the poly(A) tail, they probably increase fem-3 translation.
Using the yeast three-hybrid assay, Zhang et al. (1997) identified two proteins, called FBF-1 and FBF-2, that bind the fem-3 PME. These proteins are remarkably similar in amino acid sequence, and have overlapping functions. The gain-of-function mutations in the PME prevent the FBF proteins from binding the fem-3 3’-UTR. Moreover, inactivation of the fbf genes by RNA-mediated interference recapitulates the phenotype of these fem-3(gf) mutants — the XX animals produce sperm instead of oocytes. Thus, FBF-1 and FBF-2 repress the translation of fem-3 messages by binding the PME (Figure 5). Although fbf-1 and fbf-2 single mutants are usually wild type, recent results suggest that they are not completely redundant, but have slightly different roles in the control of sexual fate and cell proliferation (Lamont et al., 2004).
The FBF proteins are homologs of the Drosophila translational regulator Pumilio, which interacts with Nanos to inhibit translation of hunchback messages. Similarly, the C. elegans FBF proteins bind NOS-3 in the yeast two-hybrid system and in vitro (Kraemer et al., 1999). Furthermore, inactivation of NOS-3 and the two other NOS homologs by RNAi causes many XX animals to produce sperm instead of oocytes (Kraemer et al., 1999). Thus, NOS-3 is likely to act with the FBF proteins to repress translation of fem-3.
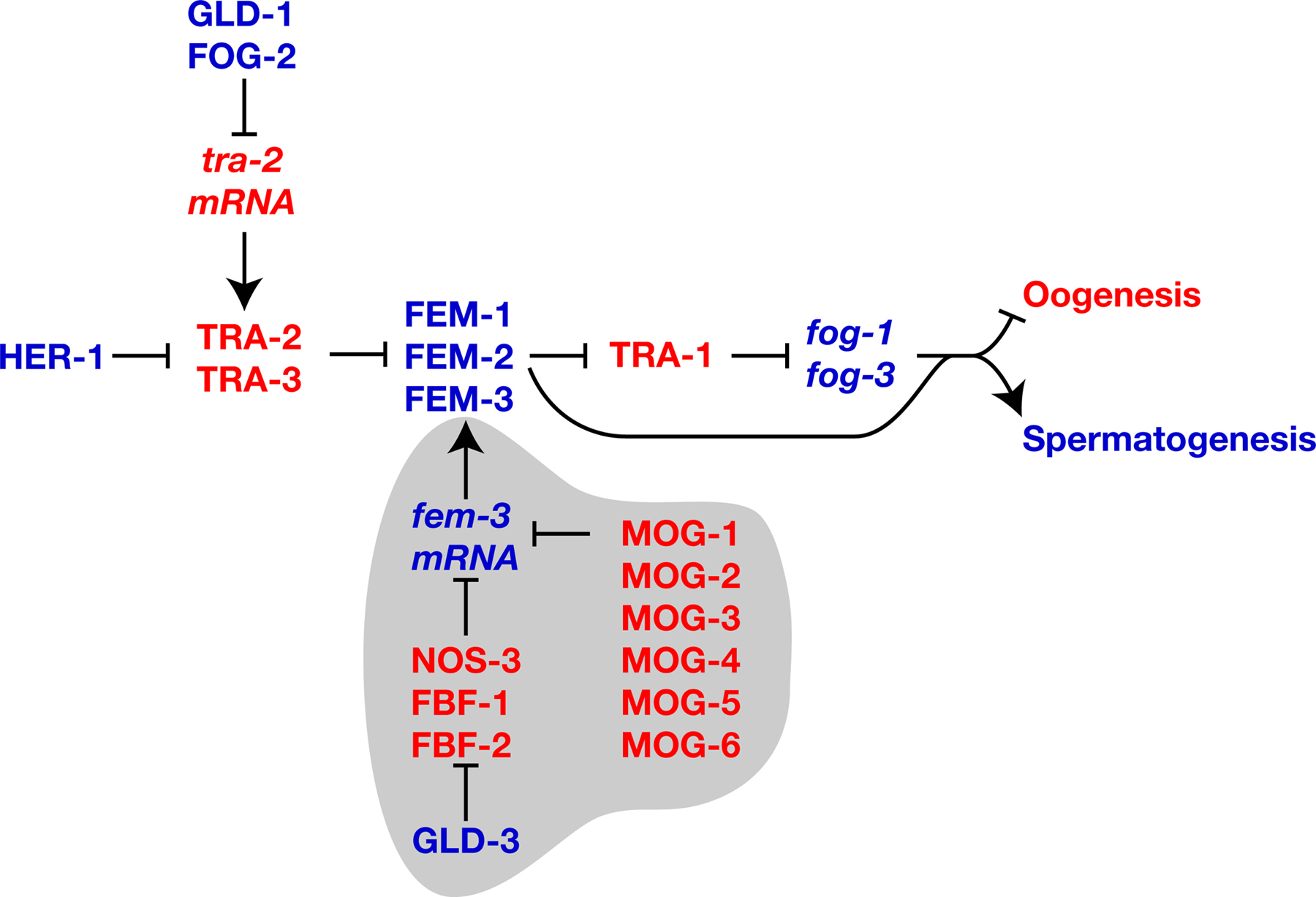 |
Figure 5. Regulation of fem-3 translation. Proteins are shown in uppercase, and genes or mRNAs in lowercase italics. Positive interactions are indicated with an arrow, and negative ones by a “--|”. Genes that promote spermatogenesis are blue, and those that promote oogenesis are red. Molecules that regulate translation of fem-3 messages are highlighted in gray.
The same yeast two-hybrid screen that identified NOS-3 also showed that the GLD-3 protein interacts with FBF-1 and FBF-2 (Eckmann et al., 2002). GLD-3 is a homolog of Bicaudal-C from Drosophila. Mutant analysis showed that GLD-3 plays several roles in germline development, one of which is the promotion of spermatogenesis — in both gld-3(null) males and hermaphrodites, some cells that should have become sperm instead develop as oocytes. This gld-3 phenotype is suppressed by mutations in the fbf genes, which implies that GLD-3 negatively regulates FBF activity (Figure 5).
The activity of fem-3 mRNA is also controlled by a second group of genes. Recessive mutations in mog-1, mog-2, mog-3, mog-4, mog-5 or mog-6 cause hermaphrodites to make sperm instead of oocytes, and also affect the size of the germ line and the viability of embryos (Graham and Kimble, 1993; Graham et al., 1993). By using reporter assays, Gallegos et al. (1998) showed that mog gene activity can also be detected in the soma, where they act through the fem-3 3’UTR to repress reporter constructs. However, mutations in the mog genes don’t cause masculinization of the soma in hermaphrodites. One possible explanation is that maternal mog activity is sufficient for normal development. Alternatively, the level of fem-3 transcripts in the soma of hermaphrodites might be outside the range that is responsive to repression by the mog genes.
Four of the mog genes have been cloned; mog-1, mog-4, and mog-5 encode DEAH-box proteins that are similar in sequence to the yeast proteins PRP16, PRP2, and PRP22, respectively (Puoti and Kimble, 1999; Puoti and Kimble, 2000), and mog-6 encodes an unusual nuclear cyclophilin (Belfiore et al., 2004). All four proteins bind the MEP-1 zinc-finger protein (Belfiore et al., 2002; Belfiore et al., 2004), which is also located in the nucleus, but has essential functions not shared by the mog genes. At this time, the manner in which the MOG complex controls target messages remains unknown.
Two other genes have similar phenotypes. First, mag-1 is the homolog of the Drosophila mago nashi gene, which regulates several aspects of germline development. In C. elegans, mag-1(RNAi) causes hermaphrodites to produce sperm instead of oocytes, and affects embryonic viability (Li et al., 2000). Although these phenotypes suggest that mag-1 functions in the same process as the mog genes, the mag-1(RNAi) phenotype is suppressed by mutations in fog-2 (Li et al., 2000), whereas the mog-1 phenotype is not (Graham and Kimble, 1993). Second, atx-2(RNAi) causes hermaphrodites to make sperm instead of oocytes, in addition to numerous other defects in germline development (Ciosk et al., 2004; Maine et al., 2004).
Although this field has reached a mature stage, in which a genetic regulatory process of great complexity has been identified and dissected into component steps, many new lines of research have opened up in the past several years and some old questions remain unsolved.
We suggested above that the crucial factor controlling whether germ cells become sperm or oocytes is the ratio of TRA-2 activity to FEM-3 activity, and described several regulatory pathways that influence these activities. However, we do not yet know why hermaphrodites make sperm early in life, and oocytes later on, since we don’t know which regulatory activities change at the appropriate time. For example, GLD-1 and FOG-2 are expressed in both larval and adult hermaphrodites, so it seems unlikely that repression of tra-2 ends when animals reach sexual maturity, thus allowing the switch to oogenesis, although it remains possible that the activity of a third factor, or a modification to either FOG-2 or GLD-1, might be responsible for the switch.
As proposed by Goodwin and Ellis (2002), three explanations might account for this puzzle. First, one of these regulatory proteins might be modified to change its activity in a manner that could not be detected by RNA or protein cytological analysis. Second, the switch might be caused by a change in the activity of a gene that has not yet been discovered. Third, the increasing size and cell number in the developing germ line might slowly change the relative activities of the tra-2 and fem-3 regulators, so that the ratio of tra-2 to fem-3 activity automatically flips during late larval development.
In theory, these two decisions might be completely independent, so that germ cells entering meiosis merely check the status of the sex-determination pathway to learn if they should begin spermatogenesis or oogenesis. However, many of the genes that act in one pathway also regulate the other. For example, the FBF proteins and GLD-1 control both processes. Furthermore, genes that control progression through the pachytene stage of meiosis also promote spermatogenesis. Are these pleiotropic effects accidental, resulting only because many genes have pleiotropic roles in development? Or do these dual activities help coordinate the overall development of the germ line? Furthermore, do these data shed any light on the developmental point at which germ cells commit to one sexual fate or the other?
The ablation of a single sheath/spermathecal precursor cell in the somatic gonad can completely feminize the germ line in XX animals (McCarter et al., 1997). This result suggests that a second somatic signal might influence germ cell fates, since her-1 is not expressed in XX animals. However, no ligand or receptor has been identified, and it is possible that these ablations exert their influence indirectly, by reducing the size of the germ line. Indeed, direct ablation of a subset of germ cells at the L2/L3 molt also completely feminizes some XX animals (McCarter et al., 1997).
There is also a potential signal transduction pathway that promotes spermatogenesis, but whose mechanism of operation remains unknown. Genes of the ERK MAP Kinase pathway are required for several different germ cell fates, including the progression of cells through the pachytene stage of meiosis I (Church et al., 1995; Hsu et al., 2002). Partial loss of function of lin-45 Raf, mek-2 MEK and mpk-1 ERK can causes hermaphrodites to make only oocytes instead of sperm. Furthermore, complete inactivation of these genes causes germ cells to arrest at the pachytene stage of meiosis and produce oocyte-specific transcripts, even in males (M. Ohmachi, M. H. Lee, E. Lambie and T. Schedl, personal communication). Thus, this pathway normally promotes spermatogenesis. However, the source of the signal, or even whether these genes act in their familiar mode as transducers of a signal, remains unknown.
Genes that regulate sexual fate in germ cells also determine if XX animals develop as females or hermaphrodites. Since new phylogenies show that hermaphroditism likely has evolved independently in C. elegans and C. briggsae (Cho et al., 2004 Kiontke et al., 2004), these nematodes provide an ideal subject for comparative evolutionary studies. In particular, we need to know how the sex-determination pathway is modified over time to create new mating systems (see The evolution of nematode sex determination).
Although we have a solid working model of sex-determination in the C. elegans germ line, the picture from Drosophila and mammals remains confusing. However, homologs of many genes that act in the C. elegans germ line are expressed in the germ cells of other animals. Do any of these genes play conserved roles in germline development, like mab-3 and doublesex do in the soma?
A major problem facing research on the C. elegans germ line is the difficulty in making reliable reporter constructs. Often, transgenic reporters are silenced in the germ line, while expression in the soma is not disrupted. This problem is at least partially caused by the ability of the germ line to shut down transcription of repetitive regions of the genome, such as are often created by transgenes. Despite the use of high-complexity DNA (Kelly et al., 1997) and particle bombardment (Praitis et al., 2001) to counter this problem, these transgenes generally take much more work to produce than similar ones in somatic tissues. New methods to simplify this process, or the identification of related nematodes with less stringent controls on expression in the germ line, could revolutionize the field.
We thank Eric Moss, Barbara Meyer, David Zarkower and Betsy Goodwin for comments, and Min Ho Lee for the photograph in Figure 4. R.E.E. was supported by American Cancer Society grant RSG-97-172-01, and T.S. was supported by NIH grant R01 GM063310.
Ahringer, J., and Kimble, J. (1991). Control of the sperm-oocyte switch in Caenorhabditis elegans hermaphrodites by the fem-3 3' untranslated region. Nature 349, 346–348. Abstract Article
Ahringer, J., Rosenquist, T.A., Lawson, D.N., and Kimble, J. (1992). The Caenorhabditis elegans sex determining gene fem-3 is regulated post-transcriptionally. EMBO J. 11, 2303–2310. Abstract
Barnes, T.M., and Hodgkin, J. (1996). The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J. 15, 4477–4484. Abstract
Barton, M.K., and Kimble, J. (1990). fog-1, a regulatory gene required for specification of spermatogenesis in the germ line of Caenorhabditis elegans. Genetics 125, 29–39. Abstract
Barton, M.K., Schedl, T.B., and Kimble, J. (1987). Gain-of-function mutations of fem-3, a sex-determination gene in Caenorhabditis elegans. Genetics 115, 107–119. Abstract
Belfiore, M., Mathies, L.D., Pugnale, P., Moulder, G., Barstead, R., Kimble, J., and Puoti, A. (2002). The MEP-1 zinc-finger protein acts with MOG DEAH box proteins to control gene expression via the fem-3 3' untranslated region in Caenorhabditis elegans. Rna 8, 725–739. Abstract Article
Belfiore, M., Pugnale, P., Saudan, Z., and Puoti, A. (2004). Roles of the C. elegans cyclophilin-like protein MOG-6 in MEP-1 binding and germline fates. Development 131, 2935–2945. Abstract Article
Chen, P., and Ellis, R.E. (2000). TRA-1A regulates transcription of fog-3, which controls germ cell fate in C. elegans. Development 127, 3119–3129. Abstract
Chen, P.J., Singal, A., Kimble, J., and Ellis, R.E. (2000). A novel member of the tob family of proteins controls sexual fate in Caenorhabditis elegans germ cells. Dev. Biol. 217, 77–90. Abstract Article
Cho, S., Jin, S.W., Cohen, A., and Ellis, R.E. (2004). A phylogeny of caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 14, 1207–1220. Abstract Article
Chu, D.S., Dawes, H.E., Lieb, J.D., Chan, R.C., Kuo, A.F., and Meyer, B.J. (2002). A molecular link between gene-specific and chromosome-wide transcriptional repression. Genes Dev. 16, 796–805. Abstract Article
Church, D.L., Guan, K.L., and Lambie, E.J. (1995). Three genes of the MAP kinase cascade, mek-2, mpk-1/sur-1 and let-60 ras, are required for meiotic cell cycle progression in Caenorhabditis elegans. Development 121, 2525–2535. Abstract
Ciosk, R., DePalma, M., and Priess, J.R. (2004). ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development 131, 4831–4841. Abstract Article
Clifford, R., Lee, M.H., Nayak, S., Ohmachi, M., Giorgini, F., and Schedl, T. (2000). FOG-2, a novel F-box containing protein, associates with the GLD-1 RNA binding protein and directs male sex determination in the C. elegans hermaphrodite germline. Development 127, 5265–5276. Abstract
Doniach, T. (1986). Activity of the sex-determining gene tra-2 is modulated to allow spermatogenesis in the C. elegans hermaphrodite. Genetics 114, 53–76. Abstract
Doniach, T., and Hodgkin, J. (1984). A sex-determining gene, fem-1, required for both male and hermaphrodite development in Caenorhabditis elegans. Dev. Biol. 106, 223–235. Abstract Article
Eckmann, C.R., Kraemer, B., Wickens, M., and Kimble, J. (2002). GLD-3, a bicaudal-C homolog that inhibits FBF to control germline sex determination in C. elegans. Dev. Cell 3, 697–710. Abstract Article
Ellis, R.E., and Kimble, J. (1995). The fog-3 gene and regulation of cell fate in the germ line of Caenorhabditis elegans. Genetics 139, 561–577. Abstract
Francis, R., Barton, M.K., Kimble, J., and Schedl, T. (1995). gld-1, a tumor suppressor gene required for oocyte development in Caenorhabditis elegans. Genetics 139, 579–606. Abstract
Francis, R., Maine, E., and Schedl, T. (1995). Analysis of the multiple roles of gld-1 in germline development: interactions with the sex determination cascade and the glp-1 signaling pathway. Genetics 139, 607–630.
Gallegos, M., Ahringer, J., Crittenden, S., and Kimble, J. (1998). Repression by the 3' UTR of fem-3, a sex-determining gene, relies on a ubiquitous mog-dependent control in Caenorhabditis elegans. EMBO J. 17, 6337–6347. Abstract Article
Goodwin, E.B., and Ellis, R.E. (2002). Turning clustering loops: sex determination in Caenorhabditis elegans. Curr. Biol. 12, R111–R120. Abstract Article
Goodwin, E.B., Hofstra, K., Hurney, C.A., Mango, S., and Kimble, J. (1997). A genetic pathway for regulation of tra-2 translation. Development 124, 749–758. Abstract
Goodwin, E.B., Okkema, P.G., Evans, T.C., and Kimble, J. (1993). Translational regulation of tra-2 by its 3' untranslated region controls sexual identity in C. elegans. Cell 75, 329–339. Abstract Article
Graham, P.L., and Kimble, J. (1993). The mog-1 gene is required for the switch from spermatogenesis to oogenesis in Caenorhabditis elegans. Genetics 133, 919–931. Abstract
Graham, P.L., Schedl, T., and Kimble, J. (1993). More mog genes that influence the switch from spermatogenesis to oogenesis in the hermaphrodite germ line of Caenorhabditis elegans. Dev. Genet. 14, 471–484. Abstract Article
Graves, L.E., Segal, S., and Goodwin, E.B. (1999). TRA-1 regulates the cellular distribution of the tra-2 mRNA in C. elegans. Nature 399, 802–805. Abstract Article
Hamaoka, B.Y., Dann, C.E., 3rd, Geisbrecht, B.V., and Leahy, D.J. (2004). Crystal structure of Caenorhabditis elegans HER-1 and characterization of the interaction between HER-1 and TRA-2A. Proc. Natl. Acad. Sci. USA 101, 11673–11678. Abstract Article
Hodgkin, J. (1980). More sex-determination mutants of Caenorhabditis elegans. Genetics 96, 649–664. Abstract
Hodgkin, J. (1986). Sex determination in the nematode C. elegans: analysis of tra-3 suppressors and characterization of fem genes. Genetics 114, 15–52. Abstract
Hodgkin, J. (1987). A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes Dev. 1, 731–745. Abstract
Hsu, V., Zobel, C.L., Lambie, E.J., Schedl, T., and Kornfeld, K. (2002). Caenorhabditis elegans lin-45 raf is essential for larval viability, fertility and the induction of vulval cell fates. Genetics 160, 481–492. Abstract
Hunter, C.P., and Wood, W.B. (1992). Evidence from mosaic analysis of the masculinizing gene her-1 for cell interactions in C. elegans sex determination. Nature 355, 551–555. Abstract Article
Jan, E., Motzny, C.K., Graves, L.E., and Goodwin, E.B. (1999). The STAR protein, GLD-1, is a translational regulator of sexual identity in Caenorhabditis elegans. EMBO J. 18, 258–269. Abstract Article
Jin, S.W., Arno, N., Cohen, A., Shah, A., Xu, Q., Chen, N., and Ellis, R.E. (2001a). In Caenorhabditis elegans, the RNA-binding domains of the cytoplasmic polyadenylation element binding protein FOG-1 are needed to regulate germ cell fates. Genetics 159, 1617–1630. Abstract Article
Jin, S.W., Kimble, J., and Ellis, R.E. (2001b). Regulation of Cell Fate in Caenorhabditis elegans by a Novel Cytoplasmic Polyadenylation Element Binding Protein. Dev. Biol. 229, 537–553. Article
Jones, A.R., Francis, R., and Schedl, T. (1996). GLD-1, a cytoplasmic protein essential for oocyte differentiation, shows stage- and sex-specific expression during Caenorhabditis elegans germline development. Dev. Biol. 180, 165–183. Abstract Article
Kawamura-Tsuzuku, J., Suzuki, T., Yoshida, Y., and Yamamoto, T. (2004). Nuclear localization of Tob is important for regulation of its antiproliferative activity. Oncogene. Abstract Article
Kelly, W.G., Xu, S., Montgomery, M.K., and Fire, A. (1997). Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146, 227–238. Abstract
Kiontke, K., Gavin, N.P., Raynes, Y., Roehrig, C., Piano, F., and Fitch, D.H. (2004). Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. USA 101, 9003–9008. Abstract Article
Kraemer, B., Crittenden, S., Gallegos, M., Moulder, G., Barstead, R., Kimble, J., and Wickens, M. (1999). NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 9, 1009–1018. Abstract Article
Kuersten, S., Segal, S.P., Verheyden, J., LaMartina, S.M., and Goodwin, E.B. (2004). NXF-2, REF-1, and REF-2 affect the choice of nuclear export pathway for tra-2 mRNA in C. elegans. Mol. Cell 14, 599–610. Abstract Article
Kuwabara, P.E. (1996). A novel regulatory mutation in the C. elegans sex determination gene tra-2 defines a candidate ligand/receptor interaction site. Development 122, 2089–2098. Abstract
Kuwabara, P.E., and Kimble, J. (1995). A predicted membrane protein, TRA-2A, directs hermaphrodite development in Caenorhabditis elegans. Development 121, 2995–3004. Abstract
Kuwabara, P.E., Okkema, P.G., and Kimble, J. (1992). tra-2 encodes a membrane protein and may mediate cell communication in the Caenorhabditis elegans sex determination pathway. Mol. Biol. Cell. 3, 461–473. Abstract
Kuwabara, P.E., Okkema, P.G., and Kimble, J. (1998). Germ-line regulation of the Caenorhabditis elegans sex-determining gene tra-2. Dev. Biol. 204, 251–262. Abstract Article
Lamont, L.B., Crittenden, S.L., Bernstein, D., Wickens, M., and Kimble, J. (2004). FBF-1 and FBF-2 Regulate the Size of the Mitotic Region in the C. elegans Germline. Dev. Cell 7, 697–707. Abstract Article
Lee, M.H., and Schedl, T. (2001). Identification of in vivo mRNA targets of GLD-1, a maxi-KH motif containing protein required for C. elegans germ cell development. Genes Dev. 15, 2408–2420. Abstract Article
Lee, M.H., and Schedl, T. (2004). Translation repression by GLD-1 protects its mRNA targets from nonsense-mediated mRNA decay in C. elegans. Genes Dev. 18, 1047–1059. Abstract Article
Li, W., Boswell, R., and Wood, W.B. (2000). mag-1, a homolog of Drosophila mago nashi, regulates hermaphrodite germ-line sex determination in Caenorhabditis elegans. Dev. Biol. 218, 172–182. Abstract Article
Luitjens, C., Gallegos, M., Kraemer, B., Kimble, J., and Wickens, M. (2000). CPEB proteins control two key steps in spermatogenesis in C. elegans. Genes Dev. 14, 2596–2609. Abstract Article
Lum, D.H., Kuwabara, P.E., Zarkower, D., and Spence, A.M. (2000). Direct protein-protein interaction between the intracellular domain of TRA-2 and the transcription factor TRA-1A modulates feminizing activity in C. elegans. Genes Dev. 14, 3153–3165. Abstract
Maekawa, M., Yamamoto, T., and Nishida, E. (2004). Regulation of subcellular localization of the antiproliferative protein Tob by its nuclear export signal and bipartite nuclear localization signal sequences. Exp. Cell Res. 295, 59–65. Abstract Article
Maine, E.M., Hansen, D., Springer, D., and Vought, V.E. (2004). Caenorhabditis elegans atx-2 Promotes Germline Proliferation and the Oocyte Fate. Genetics 168, 817–830. Abstract Article
Marin, V.A., and Evans, T.C. (2003). Translational repression of a C. elegans Notch mRNA by the STAR/KH domain protein GLD-1. Development 130, 2623–2632. Abstract Article
McCarter, J., Bartlett, B., Dang, T., and Schedl, T. (1997). Soma-germ cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev. Biol. 181, 121–143. Abstract Article
Mehra, A., Gaudet, J., Heck, L., Kuwabara, P.E., and Spence, A.M. (1999). Negative regulation of male development in Caenorhabditis elegans by a protein-protein interaction between TRA-2A and FEM-3. Genes Dev. 13, 1453–1463. Abstract
Mendez, R., and Richter, J.D. (2001). Translational control by CPEB: a means to the end. Nat. Rev. Mol. Cell Biol. 2, 521–529. Abstract Article
Mootz, D., Ho, D.M., and Hunter, C.P. (2004). The STAR/Maxi-KH domain protein GLD-1 mediates a developmental switch in the translational control of C. elegans PAL-1. Development 131, 3263–3272. Abstract Article
Nayak, S., Goree, J., and Schedl, T. (2004). fog-2 and the Evolution of Self-fertile Hermaphroditism in Caenorhabditis. PLoS Biol 3, e6. Abstract
Okkema, P.G., and Kimble, J. (1991). Molecular analysis of tra-2, a sex determining gene in C.elegans. EMBO J. 10, 171–176. Abstract
Oliver, B. (2002). Genetic control of germline sexual dimorphism in Drosophila. Int Rev Cytol 219, 1–60. Abstract
Perry, M.D., Li, W., Trent, C., Robertson, B., Fire, A., Hageman, J.M., and Wood, W.B. (1993). Molecular characterization of the her-1 gene suggests a direct role in cell signaling during Caenorhabditis elegans sex determination. Genes Dev. 7, 216–228. Abstract
Pilgrim, D., McGregor, A., Jackle, P., Johnson, T., and Hansen, D. (1995). The C. elegans sex-determining gene fem-2 encodes a putative protein phosphatase. Mol. Biol. Cell 6, 1159–1171. Abstract
Praitis, V., Casey, E., Collar, D., and Austin, J. (2001). Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157, 1217–1226. Abstract
Puoti, A., and Kimble, J. (1999). The Caenorhabditis elegans sex determination gene mog-1 encodes a member of the DEAH-Box protein family. Mol. Cell Biol. 19, 2189–2197. Abstract
Puoti, A., and Kimble, J. (2000). The hermaphrodite sperm/oocyte switch requires the Caenorhabditis elegans homologs of PRP2 and PRP22. Proc. Natl. Acad. Sci. USA 97, 3276–3281. Abstract Article
Rosenquist, T.A., and Kimble, J. (1988). Molecular cloning and transcript analysis of fem-3, a sex-determination gene in Caenorhabditis elegans. Genes Dev. 2, 606–616. Abstract
Ryder, S.P., Frater, L.A., Abramovitz, D.L., Goodwin, E.B., and Williamson, J.R. (2004). RNA target specificity of the STAR/GSG domain post-transcriptional regulatory protein GLD-1. Nat. Struct. Mol. Biol. 11, 20–28. Abstract Article
Schedl, T., Graham, P.L., Barton, M.K., and Kimble, J. (1989). Analysis of the role of tra-1 in germline sex determination in the nematode Caenorhabditis elegans. Genetics 123, 755–769. Abstract
Schedl, T., and Kimble, J. (1988). fog-2, a germ-line-specific sex determination gene required for hermaphrodite spermatogenesis in Caenorhabditis elegans. Genetics 119, 43–61. Abstract
Segal, S.P., Graves, L.E., Verheyden, J., and Goodwin, E.B. (2001). RNA-Regulated TRA-1 nuclear export controls sexual fate. Dev. Cell 1, 539–551. Abstract Article
Sokol, S.B., and Kuwabara, P.E. (2000). Proteolysis in Caenorhabditis elegans sex determination: cleavage of TRA-2A by TRA-3. Genes Dev. 14, 901–906. Abstract
Spence, A.M., Coulson, A., and Hodgkin, J. (1990). The product of fem-1, a nematode sex-determining gene, contains a motif found in cell cycle control proteins and receptors for cell-cell interactions. Cell 60, 981–990. Abstract Article
Wang, S., and Kimble, J. (2001). The TRA-1 transcription factor binds TRA-2 to regulate sexual fates in Caenorhabditis elegans. EMBO J. 20, 1363–1372. Abstract Article
Yoshida, Y., Tanaka, S., Umemori, H., Minowa, O., Usui, M., Ikematsu, N., Hosoda, E., Imamura, T., Kuno, J., Yamashita, T., et al. (2000). Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell 103, 1085–1097. Abstract Article
Zarkower, D., and Hodgkin, J. (1992). Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell 70, 237–249. Abstract
*Edited by Barbara J. Meyer. Last revised February 8, 2005. Published April 4, 2006. This chapter should be cited as: Ellis, R. and Schedl, T. Sex determination in the germ line (April 4, 2006), WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.82.1, http://www.wormbook.org.
Copyright: © 2006 Ronald Ellis and Tim Schedl. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
§To whom correspondence should be addressed. E-mail: ron.ellis@umdnj.edu or ts@genetics.wustl.edu
 All WormBook content, except where otherwise noted, is licensed under a Creative Commons Attribution License.
All WormBook content, except where otherwise noted, is licensed under a Creative Commons Attribution License.